Advanced Search
Assembly, transformation, and selection of the IL-6 library
Last updated date: Mar 2, 2021 Views: 937 Forks: 0
1. IL-6 Library Assembly
a. The assembly of the IL-6 library DNA was carried out using 14 overlapping primers covering the whole IL-6 sequence, two of which contained the NDT codon used for mutation. The primers were designed to contain the necessary homology to the pCT302 vector (highlighted in yellow), in order to clone the library into the yeast display vector (pCT302) by homologous recombination (Fig. 1).
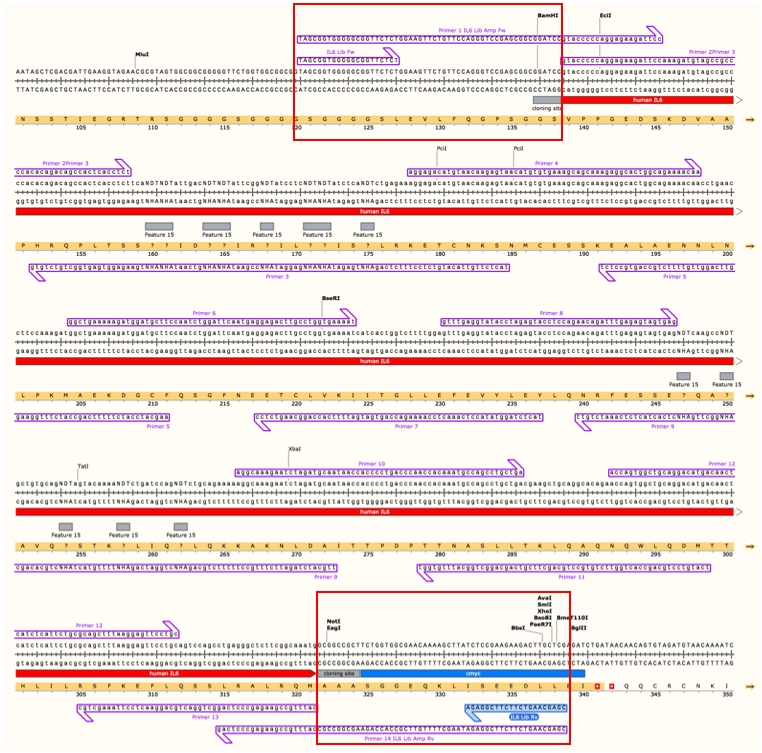
Fig 1. Primers used for the assembly of the IL-6 library. Note: primers 1 and 14 are 100% homologous to the pCT302 vector upstream and downstream of the cloning sites BamHI and NotI, respectively in order to allow for homologous recombination (red boxes). Scheme prepared with SnapGene.
b. The primers used were:
Primer 1 (IL-6 library amplification Fw): TAGCGGTGGGGGCGGTTCTCTGGAAGTTCTGTTCCAGGGTCCGAGCGGCGGATCCGTACCCCCAGGAGAAGATTCC
Primer 2: GTACCCCCAGGAGAAGATTCCAAAGATGTAGCCGCCCCACACAGACAGCCACTCACCTCT
Primer 3: TACTCTTGTTACATGTCTCCTTTCTCAGAHNTGAGATAHNAHNGAGGATAHNCCGAATAHNAHNGTCAATAHNAHNTGAAGAGGTGAGTGGCTGTCTGTG
Primer 4: AGGAGACATGTAACAAGAGTAACATGTGTGAAAGCAGCAAAGAGGCACTGGCAGAAAACAA
Primer 5: AAGCATCCATCTTTTTCAGCCACTTTTGGAAGGTTCAGGTTGTTTTCTGCCAGTGCCTCT
Primer 6: GGCTGAAAAAGATGGATGCTTCCAATCTGGATTCAATGAGGAGACTTGCCTGGTGAAAAT
Primer 7: TACTCTAGGTATACCTCAAACTCCAAAAGACCAGTGATGATTTTCACCAGGCAAGTCTCC
Primer 8: GTTTGAGGTATACCTAGAGTACCTCCAGAACAGATTTGAGAGTAGTGAG
- Primer 9: TTGCATCTAGATTCTTTGCCTTTTTCTGCAGAHNCTGCAGAHNTTTTGTACTAHNCTGCACAGCAHNGGCTTGAHNCTCACTACTCTCAAATCTGTT
- Primer 10: AGGCAAAGAATCTAGATGCAATAACCACCCCTGACCCAACCACAAATGCCAGCCTGCTGA
- Primer 11: TCATGTCCTGCAGCCACTGGTTCTGTGCCTGCAGCTTCGTCAGCAGGCTGGCATTTGTGG
- Primer 12: ACCAGTGGCTGCAGGACATGACAACTCATCTCATTCTGCGCAGCTTTAAGGAGTTCCTGC
Primer 13: CATTTGCCGAAGAGCCCTCAGGCTGGACTGCAGGAACTCCTTAAAGCTGC
- Primer 14 (IL-6 library amplification Rv):: CGAGCAAGTCTTCTTCGGAGATAAGCTTTTGTTCGCCACCAGAAGCGGCCGCCATTTGCCGAAGAGCCCTCAG
c. NDT codons allow for the mutation of selected amino acids by replacing those amino acids with Glycine (G), Valine (V), Leucine (L), Isoleucine (I), Cysteine (C), Serine (S), Arginine (R), Histidine (H), Aspartic acid (D), Asparagine (N), Phenylalanine (F) and Tyrosine (Y).
d. The following amino acids from IL-6 were chosen to randomize: D9, E22, R23, K26, Q27, Y30, D33, G34, A37, E109, R112, M116, V120, F124.
e. In order to assemble the IL-6 library, mix 5 μL of each primer resuspended at 100μM (i.e. overlapping primers covering the whole sequence of IL-6 including NDT primers designed to contain the mutations of interest, see above) with 10 μL of each of the IL-6 library amplification forward (Fw) and IL-6 library amplification reverse (Rv) primers and do a 1:2 serial dilution 12 times (i.e. 100μM, 50μM, 25μM, 12.5μM, 6.25μM, 3.125μM, etc). PCR Conditions used were: 98°C for 1 minute, 35 cycles of: 98°C for 10 seconds/55°C for 30 seconds/72°C for 1 minute, then 72°C for 10 minutes (Fig.2).
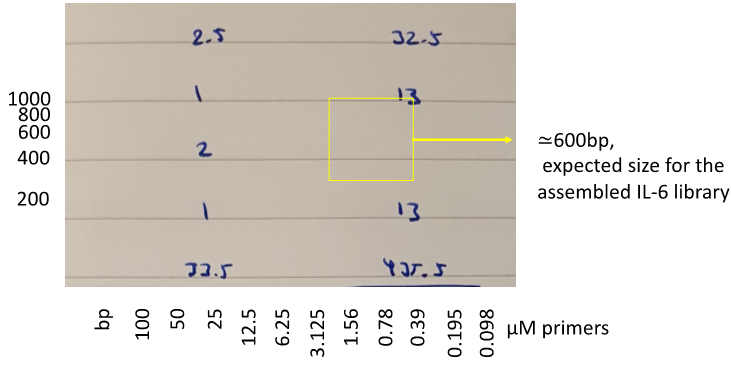
Figure 2. Example of the expected results after the PCR for the library assembly. Expected outcome highlighted with a yellow box.
f. Once generated, the IL-6 library was further amplified in order to obtain around 50-100μg of DNA (i.e. normally this would require about fifty 50μL PCR reactions) (Fig.3) using the primers:
IL-6 Lib Fw: 5’- TAGCGGTGGGGGCGGTTCTCT−3’ and
IL-6 Lib Rv: 5’- CGAGCAAGTCTTCTTCGGAGA −3’.
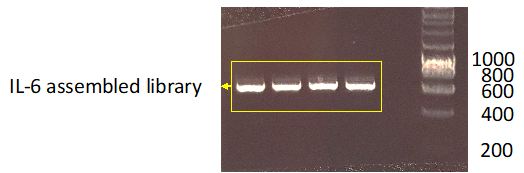
Figure 3. Example of the expected results after the library amplification by PCR.
2. EBY100 yeast Electroporation
Day 1. Thaw a vial of EBY100 yeasts on ice and plate the cells on a YPD plate with ColiRollers plating beads, and allow cells to grow at 30ºC over the weekend.
Day 2. Inoculate 5 mL of YPD media supplemented with chloramphenicol (35μg/mL, final concentration) to avoid bacterial contamination with a swipe of EBY100 colonies and grow the cells ON at 30°C and 250rpm.
Day 3.
Measure OD600 of the ON culture. Note: OD600=1 equals to 107 yeast/mL.
Dilute the yeast culture in 100 mL of YPD medium at an OD600 of 0.2-0.4. Allow the culture to grow in a baffled flask at 30°C and 250rpm until it reaches an OD600=1.3-1.5. (4-6 hours)
Then, add 1 mL of Tris-DTT and shake the cells again at 30°C and 200rpm for 15 minutes. Note: DTT is required to reduce the yeast surface facilitating the DNA entry and increasing transformation efficiency.
Divide into two 50ml falcon and centrifuge for 3 minutes at 2500g and 4°C.
Remove the supernatant and resuspend each pellet in 25 mL E buffer.
Centrifuge for 3 minutes at 2500g and 4°C.
Pre-heat 50 mL of YPD media to 30ºC at this stage
Resuspend each pellet in 1 mL E buffer and transfer each one into microcentrifuge tubes.
Centrifuge for 3 minutes at 2500g and 4°C.
Resuspend both pellets in 300 µL E buffer + insert and vector in a 5:1 ratio (at least 25µg:5µg). Note: Do not exceed 300 uL of insert-vector mix to avoid too much dilution of the yeast.
Aliquot 50 µL volumes into each chilled electroporation cuvettes. Around 18 cuvettes.
Electroporate with voltage 500V (LV) capacitance 25µF, resistance none. Note: Time constant should be between 50-10ms. Avoid touching metal electrodes on electroporation cuvettes.
Quickly add 1 mL of 30°C-heated YPD medium to the cuvette and transfer to a baffled flask, then wash the cuvette with another mL of medium adding this to the flask. Note: Tilt the cuvettes to get out maximum of the yeast suspension
Add media up to 50 mL if necessary. Note: Top up with the rest of the unused pre-warmed YPD media.
Put the flask in a 30°C incubator for 1h at 250rpm.
Centrifuge the yeasts for 3 minutes at 3000g and 4°C and discard the supernatant.
Resuspend in 10 mL of SDCAA medium (1.3x107 cell/ml).
Keep 10 µL to add to 990 µL SDCAA medium (104) then perform 3 1:10 serial dilutions in a final volume of 1mL (105, 106, 107) and plate 100 µL of each one on a SDCAA plate. Incubate at 30°C for 3 days. Note: This will allow us to estimate the size of our library. For example, if we get 100 colonies in our 106 plate and 8 colonies in our 107 plate the size of our library will be between 1x108 and 8x107.
Put the remaining yeast culture in 500 mL SDCAA in a glass baffled Fernbach flask and incubate at 30°C and 250rpm. Add chloramphenicol at 35μg/mL.
Day 4. After 24h measure the OD600 and put 1x1010 yeast in 500 mL fresh SDCAA to grow for 24h at 30°C (add chloramphenicol at 35μg/mL). Note: When the yeasts are growing properly the OD600 should be at least 4-5 and the culture looks milky.
Day 5.
After 24h measure the OD600 and put 1x1010 yeast in 500 mL fresh SGCAA and grow for 48-72h at 20°C to induce protein expression and start with the library selection.
Take 1x1010 yeast and put them in 500 mL of SCDAA low dextrose for 24-48h at 30°C before freezing the cells. Note: This step is required in order to keep aliquots of your original library before starting the selection, allowing you to go back to repeat the selection if required or under different experimental conditions.
3. IL-6 Library Selection
1st round:
Measure the OD600 and take 1x1010 cells.
Wash with PBE and centrifuge for 3 minutes at 2500g two times discarding the supernatant.
Resuspend the yeast in 5ml of PBE + 100µl Miltenyi Streptavidin Microbeads.
Rotate for 1h at 4°C.
Spin down the cells for 5 minutes at 2500g and discard the supernatant.
Set up the magnetic LS column.
1) Place the column in the magnetic rack in the cold room. All the subsequent steps will be carried out at 4°C.
2) Equilibrate the column with 5 mL PBE.
3) Prepare one tube to collect the flow-through.
Resuspend the pellet in 3 mL PBE and add it to the LS column.
Wash the column with 3 mL PBE three times.
Collect the entire flow-through. Note: These steps allow the removal of all the yeast that bind to streptavidin in the absence of biotinylated protein, therefore, the flow-through contains the yeast of interest.
Pellet the cells and remove the supernatant.
Mix 250 µL streptavidin-beads with your biotinylated protein at a final concentration of 400nM and leave for 10-15 min at RT. Note: The biotinylated protein will form a tetramer with increased avidity for the target protein expressed on the library.
Resuspend the pellet in 4.7 mL PBE and then add 250 µL streptavidin-beads conjugated to your protein.
Rotate for 2h at 4°C.
Pellet the cells, discard the supernatant, and resuspend in 3 mL PBE.
Equilibrate a new LS column with 5 mL PBE.
Add the cells to the LS column.
Wash the column with 3 mL PBE three times.
Remove the column from the magnet and elute with 5 mL PBE using the plunger, collecting the eluate in tube. You have to collect all the cells binding to the protein of your interest.
Pellet the eluate and remove the supernatant.
Wash with SDCAA.
Resuspend the pellet in 4 mL SDCAA and incubate at 30°C ON.
Next day measure the OD600, dilute to OD600 = 1 in 4 mL SGCAA. Induce protein expression by incubating at 20°C for 48h.
2nd round: c-myc expression. This round aims to select yeast cells expressing full-length variants of IL-6. For that, it uses the c-myc tag present at the C-ter of IL-6. Note: If at any moment of the selection the percentage of c-myc positive cells is lower than 20% you have to start again from the previous round and repeat the selection.
i. Collect 5 µL yeast to check c-myc expression, as follows:
1) Wash with 1 mL PBE, then centrifuge for 3 minutes at 2500g.
2) Resuspend in 100 µL of PBE and divide in 2 aliquots of 50 µL each.
3) Add 1 µL anti-myc-647 to one sample. Keep one of the two aliquots as unstained control.
4) Incubate for 30 minutes at 4°C covered with foil.
5) Wash with PBE and pellet as before.
6) Resuspend in 200 µL PBE and analyze on the flow cytometer. Note: You should obtain a big negative population and a smaller positive one.
ii. Measure the OD600 and take 108 yeast.
iii. Pellet the yeast and discard the supernatant.
iv. Resuspend the pellet in 495 mL of PBE + 5 mL of anti-cmyc-647 Ab and transfer to a microcentrifuge tube.
v. Rotate for 2h at 4°C covered with foil in order to protect the fluorochrome.
vi. Add 1 mL PBE to the microcentrifuge tube, pellet the cells by centrifugation for 3 minutes at 4000g, and remove the supernatant.
vii. Resuspend the pellet in 950 mL PBE + 50mL of anti-647 beads.
viii. Rotate for 20 minutes at 4°C covered with foil.
ix. Pellet the cells and remove the supernatant.
x. Equilibrate an LS column with 5 mL PBE.
xi. Resuspend the pellet in 3 mL PBE, keep 5 mL for analysis on the flow cytometer, and gently add the remaining sample to the column down the wall of the column.
xii. Wash the column with 3 mL PBE three times and collect the flow-through.
xiii. Remove the column from the magnet and elute with 5 mL PBE into a culture tube using the plunger.
xiv. Analyze at the flow cytometer the unstained, pre-sort, flow-through and post-sort samples.
xv. Pellet the eluate, discard the supernatant, resuspend the pellet in 3 mL SDCAA and grow the cells ON at 30°C. Note: Remove all of the PBE or the yeast growth will be slower.
xvi. Next day measure the OD600, dilute to OD600=1 and induce expression in 3 mL SGCAA at 20°C for 48h.
c. 3rd round: 100nM tetramer = 100nM streptavidin + 400nM gp130
Collect 3 µL yeast to check myc expression.
Measure the OD600 and take 108 yeast. Keep 5 mL for analysis on the flow cytometer.
Pellet the yeast and discard the supernatant.
Prepare 500 mL PBE with 100nM streptavidin647 and 400nM gp130-biotinylated and incubate for 10 minutes at RT.
Wash yeast with PBE two times. Keep 5 mL for analysis on the flow cytometer.
Resuspend yeast in 500 mL containing the tetramer from step iv.
Rotate for 2h at 4°C covered with foil.
Add 1 mL PBE to the microcentrifuge tube, and pellet the cells for 3 minutes at 4000g and remove the supernatant.
Resuspend the pellet in 950 mL PBE + 50 mL of anti-647 beads.
Rotate for 20 minutes at 4°C covered with foil.
Pellet cells, discard the supernatant, and resuspend in 3 mL PBE. Keep 5 mL for analysis on the flow cytometer.
Equilibrate a new LS column with 5 mL PBE.
Wash the column with 3 mL PBE X3 times.
Collect the flow-through. Keep 5 mL of the flow-through for the control at the flow cytometer.
Remove the column from the magnet and elute with 5 mL PBE using the plunger, collecting the eluate in tube. You have to collect all the cells binding to the protein of your interest.
Analyze at the flow cytometer the unstained, pre-sort, flow-through and post-sort samples.
Pellet the eluate, discard the supernatant, resuspend the pellet in 3 mL SDCAA and grow the cells ON at 30°C.
Next day measure the OD600, dilute to OD600=1 and induce expression in 3 mL SGCAA at 20°C for 48h.
d. 4th round of selection: 1μM gp130
Collect 3 µL yeast to check myc expression.
Measure the OD600 and take 108 yeast. Keep 5 mL for analysis on the flow cytometer.
Transfer yeast to a 15 mL tube, add 6 mL PBE, spin down at 2500g and discard the supernatant.
Wash yeast with PBE two times. Keep 5 mL for analysis on the flow cytometer.
Resuspend the yeast in 300 uL PBE, transfer to a microcentrifuge tube and incubate with 1uM of biotinylated-GP130.
Rotate for 1h at 4°C.
Add 1 mL PBE to the Eppendorf, and pellet the cells for 3 minutes at 4000g. Remove the supernatant.
Wash once with 1 mL of PBE
Resuspend the pellet in 500 mL PBE + 100nM streptavidin-647 and rotate for 15 minutes at 4°C covered with foil.
Wash 1x with 1 mL of PBE
Resuspend the pellet in 950 mL PBE + 50 mL of anti-647 beads.
Rotate for 20 minutes at 4°C covered with foil.
Pellet cells, discard the supernatant, and resuspend in 3 mL PBE. Keep 5 mL for analysis on the flow cytometer.
Equilibrate a new LS column with 5 mL PBE.
Add the 3 mL of yeast suspension to the LS-column. Collect the flow-through
Wash the column with 3 mL PBE three times.
Collect the flow-through. Keep 5 mL of the flow-through for analysis on the flow cytometer.
Remove the column from the magnet and elute with 5 mL PBE using the plunger, collecting the eluate in tube. You have to collect all the cells binding to the protein of your interest.
Analyze at the flow cytometer the unstained, pre-sort, flow-through and post-sort samples.
Pellet the eluate, discard the supernatant, resuspend the pellet in 3 mL SDCAA and grow the cells ON at 30°C.
Next day measure the OD600, dilute to OD600=1 and induce expression in 3 mL SGCAA at 20°C for 48h.
e. 5th and subsequent rounds of selection. The next rounds of selections aim to increase the binding affinity of the library by purifying variants with increased affinity for the target protein (gp130 in our case). For that reason, in the next rounds of selection we will continue decreasing the concentration of gp130 (e.g. 5th round 200nM, 6th round 50nM, and so on) until we achieve our target affinity.
Material:
E buffer: 1.2 g Tris base, 92.4g sucrose, 0.2 g MgCl2 in 1L of dH2O (use high quality H2O in order to keep proper conductivity) and adjust pH to 7.5. Filter the solution and keep it at 4°C.
Tris-DTT: 0.39g 1,4-dithiothreitol (DTT) in 1 mL 1 M Tris, pH 8.0 and sterilize by filtration. Always make fresh buffer.
PBE: PBS containing 2mM EDTA and 0.5% BSA
YPD media: 10 g yeast extract, 20g peptone, 20g dextrose (D-glucose) in 1 L of dH2O. Autoclave.
YPD plates: as the YPD media but add 15g Bacto-Agar. Add ddH2O to final volume and autoclave.
SDCAA media: 20g dextrose, 6.7g Difco yeast nitrogen base, 5g Bacto aminoacids, 14.7g sodium citrate and 4.29g citric acid in ddH2O to a volume of a liter. Filter sterilize and store at +4ºC.
SGCAA media: as the SDCAA media but using 20g of Galactose instead of dextrose.
Freezing buffer: 0.335g Yeast Nitrogen Base, 1 mL glycerol, 50 mL H2O. Filter to sterilize and store at 4 °C.
- Martinez-Fabregas, J, Pohler, E and Moraga, I(2021). Assembly, transformation, and selection of the IL-6 library. Bio-protocol Preprint. bio-protocol.org/prep897.
- Martinez-Fabregas, J., Wilmes, S., Wang, L., Hafer, M., Pohler, E., Lokau, J., Garbers, C., Cozzani, A., Fyfe, P. K., Piehler, J., Kazemian, M., Mitra, S. and Moraga, I.(2019). Kinetics of cytokine receptor trafficking determine signaling and functional selectivity. eLife. DOI: 10.7554/eLife.49314
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
