Advanced Search
Quantifying γδT-cell activation in vivo and in vitro
Last updated date: Nov 12, 2025 Views: 1174 Forks: 0
Abstract
The skin plays an important role in protecting the body from pathogens and chemicals in the external environment. Upon injury, a healing program is rapidly initiated and involves extensive intercellular communication to restore tissue homeostasis. The deregulation of this crosstalk can lead to abnormal healing processes and is the foundation of many skin diseases. A relatively overlooked cell type that nevertheless plays critical roles in skin homeostasis, wound repair, and disease is dendritic epidermal T cells (DETCs). Given their varied roles in both physiological and pathological scenarios interest in the regulation and function of DETCs has substantially increased. Moreover, their ability to regulate other immune cells has garnered substantial attention for their potential role as immunomodulators and in immunotherapies. In this article we describe a protocol to isolate and culture DETCs and their in vivo analysis within the skin. These approaches will facilitate the investigation of their crosstalk with other cutaneous cells and the mechanisms by which they influence the status of the skin.
Keywords:
Skin, Wound healing, immune cells, dendritic epidermal cells, Proliferation, Histology, Immunohistochemistry
Background:
The skin is composed of multiple cell types which include keratinocytes, fibroblasts and various immune cells, that work together to provide the physical and immune barrier against the external environment. One of the skin resident immune cells, dendritic epidermal T cells (DETCs), has been shown to play important roles in tissue homeostasis, repair and pathophysiology. In the epidermis, DETCs exhibit a dendritic morphology that allows each cell to stay in physical contact with other epidermal cells such as keratinocytes and Langerhan cells (Jameson & Havran, 2007). This intercellular communication between keratinocytes and Langerhan cells is mediated by factors which activate DETCs, which in turn modulates tissue homeostasis. For example, during wound healing, damaged and stressed keratinocytes express various antigens that are recognized by DETCs resulting in their activation and release of various cytokines such as keratinocyte growth factor-1 and 2, insulin growth factor-1, and interleukin-2. In addition DETCs also secrete fibroblast growth factor 9, which mediates hair neogenesis during wound healing (Gay et al., 2013). Recently, we have uncovered a novel role of skin resident DETCs in regulating hair follicle stem cell activity in wounded skin (Lee et al., 2017). In addition DETCs have also been shown to play an important role in maintaining the epidermis in unwounded skin as mice lacking DETCs exhibit higher levels of epidermal apoptosis (Sharp, Jameson, Cauvi, & Havran, 2005). In this protocol, we provide a detailed method of the isolation and culturing of DETCs to investigate the effect of various soluble factors and cell-cell contacts on DETC activation and the downstream consequence on other cutaneous cells. We also describe a method of in vivo analysis of DETCs, which will help in understanding the intercellular communication of DETCs at a tissue level in both physiological and pathological conditions.
Reagent and recipe:
1. C57Bl6 mice (Jackson laboratories)
2. 70% Ethanol.
3. Fetal Bovine Serum (Gibco catalog number 10270106)
4. Phosphate buffer solution (PBS)(cshprotocols)
5. Trypsin solution. (Sigma Aldrich catalog number T4049)
6. E media without calcium (Nowak & Fuchs, 2009)
7. 7-AAD. (ThermoFisher Scientific catalog number A1310)
8. FITC isotype control (ThermoFisher Scientific catalog number GM4992)
9. PE-Cy7 isotype control (ThermoFisher Scientific catalog number 25-4714 - 80)
10. PE isotype control (ThermoFisher Scientific catalog number 12-4714-42)
11. FITC anti-Vγ3 TCR (ThermoFisher Scientific catalog number MHGD01)
12. PE-Cy7 anti-CD3ε (ThermoFisher Scientific catalog number 25-0038-42)
13. PE anti- γδ TCR (ThermoFisher Scientific catalog number 12-9959-42)
14. Concanavalin A (Sigma Aldrich catalog number C5275)
15. Recombinant IL-2(Promo cell catalog number-61241)
16. Glutamine (Sigma Aldrich, catalog number G8540)
17. HEPES (Sigma Aldrich, catalog number H3375-25G)
18. Sodium pyruvate (HImedia, catalog number PCT0503)
19. NEAA (Merck M catalog number 7145 M7145)
20. Penicillin (Sigma Aldrich catalog number 1502701)
21. streptomycin (Sigma Aldrich catalog S9137-25G)
22. β-Mercapto ethanol (ME) (Sigma Aldrich catalog number M6250)
23. Recombinant human IL2 (Promo cell catalog number-61241)
24. Gentamicin (Thermo Fisher Scientific, catalog number 15750078)
25. Mouse IL-17 Quantikine ELISA Kit (R&D systems catalog number M1700)
26. FGF7 (Sigma Aldrich, catalog number RAB0188)
27. TNFα (Thermo scientific catalog number KHC3011)
28. IFN-γ (Thermo scientific catalog number RAB0223)
29. IL23 (ThermoFisher Scientific catalog number PHC9321.)
30. Recombinant IL-1beta (10 ng/ml), (R&D systems, 201-LB-005/CF)
31. DMEM/F-12, powder (Gibco, catalog number12500062)
32. Sodium bicarbonate (Gibco catalog number S5761)
33. Cholera toxin (Sigma Aldrich catalog numberC8052-.5MG)
34. Hydrocortisone (Sigma Aldrich catalog number H0888)
35. Autoclaved milli-Q distilled water.
36. Hydrochloric acid (Sigma Alddrich catalog number 320331-500ML)
37. Anti-CD3 (1 μg/ml) (Abcam,catalog number 5690)
38. Trypan blue. (ThermoFisher Scientific, 15250061)
39. Anti-JAML (Abcam, catalog number 67843)
40. WST-1 reagent (Merck, catalog number 5015944001)
41. MTT reagent. (Merck, catalog number CT01-5)
42. OCT medium (Thermo scientific, catalog number 23-730-571)
43. 16% paraformaldehyde (Fisher scientific, catalog number 50-980-487)
44. Triton X-100 (Thermo Scientific™ PI28313)
45. Keratin 5 (Abcam, ab52635)
46. Anti-Ki67 (Abcam, catalog number ab16667)
47. Anti γδTCR antibody (eBioscience, catalog number 12-5711-82)
48. Goat anti Rabbit Alexa fluor 488 (Molecular probes, catalog number A-11008)
49. Goat anti-chicken Alexa fluor 647 (Molecular probes, catalog number A-21449)
50. DAPI (Abcam, catalog number ab228549)
51. Vectashield (Vector laboratories, catalog number H-1500)
Equipment
1. Scissors (Fisher scientific catalog number 08-951-20)
2. Fine forceps. (Fisher scientific catalog number NC9924848)
3. 10ml serological pipette (Stem cell technologies catalog number 38004)
4. 70µm cell strainer. (Corning, catalog number 431751)
5. 10cm petri dish. (Eppendorf catalog number 30702118)
6. Cell culture incubator (Eppendorf, Model number Eppendorf™ Galaxy™ 170
7. Aspirator
8. FACS tubes (Stem cell technologies catalog 38007)
9. 50ml Falcon tube (Thermo fisher, Item code:10788561)
10. 15 ml falcon tubes. (Stem cell technologies catalog 05860)
11. FACS Aria (BD FACSAria™ III sorter)
12. Centrifuge (Eppendorf Model number 5702)
13. Ultra-low attachment culture plates (Corning catalog number CLS3471-24EA)
14. 96 well dish (Eppendorf EP0030730011-80EA)
15. Spectrophotometer. (Thermo Scientific™ GENESYS™ 20 Visible Spectrophotometer)
16. Cell culture incubator (Eppendorf, Model number Eppendorf™ Galaxy™ 170 S)
17. cell counter (Thermo fisher scientific)
18. Cryostat (Leica CM1950)
19. -80 °C freezer (Thermo scientific, Forma Ultra-Low Temperature Upright DD Freezer)
20. Kimwipes. (Kimberly-Clark Kimtech Science Kimwipes Wipers)
21. Hydrophobic pen (Merck catalog number Z377821-1EA)
22. Humidifying chamber
23. Compound Binocular Microscope (Celestron Labs, Model: CB2000CF)
24. Fluorescent microscope (Olympus, IX73), Confocal microscope FV 3000 5 laser (IEC60825-1:2007)]
25. Image J software
26. Graphpad Prism
Recipe:
1. RPMI media
Add the following components in 600ml sterile distilled water.
10% FCS, 2 mM Glutamine, 25 mM HEPES, 1 mM Sodium pyruvate, 100ug NEAA, 100U penicillin,100uG streptomycin, 50 uM 2ME, 20U/ml recombinant human IL2
Make up the volume up to 1000ml. Filter sterilize the media before use.
2. FACS Staining Buffer:
Add 1ml of chelated Fetal Bovine Serum, 1X Penicillin streptomycin,50ug/ml amicin), PBS to make the final volume to 50ml.
3. PBS with antibiotics
Add 100U penicillin and 100uG streptomycin to 10X PBS and makeup volume up to 1L.
4. E media without calcium (Nowak & Fuchs, 2009)
Preparation of E Media Without Calcium for Epidermal Cells
In a 6 L Erlenmeyer flask, combine six packets of Gibco Invitrogen customized
DMEM: F12 (3:1) without calcium with glass distilled H2O to reach a final volume of 5.5 L.
i. Add 18.42 g of sodium bicarbonate, 2.85 g of L-glutamine, and 60 mL of 100× penicillin- streptomycin solution.
ii. Adjust pH to 7.2 using 10 N HCl and adjust the volume to 6 L with H2O.
iii. Apply compressed CO2 to the media for 15 min. The media should reach an amber color.
iv. Combine stock additives with chelated FBS prepared in 3.1.2 by adding 75 ml of 100× cocktail, 750 µl cholera toxin and 750 µl hydrocortisone to 1 L of chelated FBS.
v. Produce final 15% FBS media in 1 L batches by combining 850 ml of the DMEM: F12 media base from step 4 with 150 ml of the supplemented chelated FBS from step 5 and sterilize using a Nalgene bottle top filter.
vi. Media can be stored in 250 or 500 mL bottles at −20 °C.
vii. Note that calcium must be added to media used for cell culture, but must be omitted from media used in the cell preparation for FACS.
5. Triton X-100
Add 2ml of 100% Triton X 100 in 100ml 10X PBS. Make up the volume up to 1L.
6. Blocking solution for permeabilization
Add 0.5ml of goat serum (or serum originating from species same as the secondary fluorescent conjugated antibody) in 10ml of 0.1% triton X 100 solution diluted in PBS.
7. 4% Paraformaldehyde (PFA)
Dilute the 16% paraformaldehyde PBS by adding 12.5 ml of 16% of PFA to 37.5 ml of PBS.
Methods:
Isolation, maintenance, and proliferation of DETCs
Given the important roles of DETCs, methods to investigate the regulation and function of these cells are required. This is facilitated by the ability to isolate and establish primary cultures of DETCs and reconstitute their intercellular crosstalk with different cells in vitro.
Isolation and culturing of DETCs
1. Isolation of DETCs from the skin via FACS (Kashem & Kaplan, 2018)
i) Euthanize C57/Bl6 pups of postnatal day 0 to 5 via decapitation or an approved method of the Institutional Animal Ethics Committee. 3 to 4 pups are required for a 3.5 cm dish of cultured DETCs.
ii) Clean the surface of the pup using 70% ethanol to decrease the chances of microbial contamination.
iii) Remove the limbs and tail of each pup as close to the core body as possible using sharp scissors. Insert the scissors through the hole made by the removal of the tail and cut the skin along the dorsal midline of the body all the way to the neck.
iv) Using forceps grasp the skin and peel the whole skin off of the body taking care not to tear the skin into pieces. Rinse the peeled skin by placing it in a tube with 10ml of sterile PBS containing 2X antibiotics (see reagent 1) for 10 minutes. Then remove excess PBS by blotting the skin on a tissue.
v) In a new 10 cm petri dish containing 10ml of 0.25% trypsin, place the skin with dermis side down making contact with the solution. Avoid submerging the epidermis in the trypsin solution to prevent over digestion of the epidermis. Spread out the curled edges using fine forceps to maximize contact of the entire dermis with the trypsin solution.
vi) Incubate the skin in trypsin for one hour at 37 °C or overnight at 4 °C.
vii) Separate the epidermis from the dermis using fine forceps. The separated epidermis will appear as a thin opaque sheet. The dermis can be used to isolate dermal cells such as fibroblasts (Kashem & Kaplan, 2018). To view a detailed protocol and video of epidermal cells isolation refer to Li et al., 2017. Cut the epidermis into small pieces with scissors. Transfer the epidermis and trypsin mixture to a new 50ml tube.
viii) Using a 10ml serological pipette, repeatedly pipette the mixture of epidermis and trypsin up and down to facilitate the dissociation of the tissue into individual cells. The serological pipette might get blocked due to clumps of tissue. Tap the pipette to remove the clumps from the pipette. Keep on pipetting until it becomes easy to pipette up and down without tapping.
ix) Pass the cell suspension through a 70µm cell strainer into a new 50 ml tube. Also, pass 5ml of E media without calcium (see reagent 2) through the cell strainer to remove any cells that are trapped in the strained, which would also inactivate the trypsin
x) Centrifuge the cell suspension at 250g for 10 minutes in a swing bucket rotor at 4 °C.
xi) Remove the supernatant carefully using an aspirator. Wash the cells once with PBS by resuspending the cell pellet in 5 ml of PBS and centrifugation at 250g for 5 minutes each.
xii) Resuspend the cells in 1ml of staining buffer (see reagent 3) for FACS sorting.
For Staining Buffer use chelated fetal bovine serum (see reagent 4) since the presence of calcium might lead to formation of cell clumps.
xiii) Count the number of viable cells using trypan blue staining
xiv) To prevent the nonspecific binding of antibodies, incubate the cells with anti CD16/CD32 (1µg/million cells) for 5 minutes at 4 °C. CD16/CD32 incubation prevents nonspecific binding of immunoglobulins to FcγIII, FcγII and possibly FcγI receptors.
xv) From this epidermal cell suspension make five aliquots of 50 µl each in five different FACS tubes. Add 50 µL of staining buffer to each 50 µL aliquot to match the volumes listed in Table 1 and label samples as indicated.
At least 100,000 cells are required to calibrate the flow cytometer.
Table 1
Summary of control and analyte samples required for FACS analysis
To calibrate the flow cytometer various controls are required: unstained control (#1), and single stained and isotype controls (#2-5). For samples which are to be used to isolate DETCs (#6/analyte sample) cells are stained with all antibodies and resuspended in a staining buffer with 5 µL of 7AAD (Fluorochrome used to stain non-viable cells). Samples used for isolating DETCs might need to be diluted such that the event rate on a FACS machine is between 1000-4000 events/sec (It is ideal to start with higher concentration and dilute it later based on the event rate).
Sample | Sample type | Volume | Antibody dilution | Incubation time(in dark) |
1.Unstained | Control | 100ul | - | 30 mins |
2.7-AAD | Control | 100ul | 0.5ul | 30 mins |
3.FITC isotype control | Control | 100ul | 1:10 | 30 mins |
4. PE-Cy7 isotype control | Control | 100ul | 1:50 | 30 mins |
5. PE isotype control | Control | 100ul | 1:200 | 30 mins |
6. Analysis | Analyte | 500ul | FITC anti-Vγ3 TCR(1:10) PE-Cy7 anti-CD3ε(1:50) PE anti- γδ TCR (1:200) 7-AAD (5ul) | 30 mins |
xvi) After incubating the cells for 30 minutes in the dark, pellet the cells by centrifuging the tubes in a swing bucket rotor at 250g for 5 minutes at 4 °C. Remove the supernatant carefully with an aspirator. Wash the cell pellet three times by re-suspending it in 1ml of staining buffer and centrifuging at 250 g for 5 minutes.
xvii) DETCs can then be isolated using FACS explained in detail in Badarinath et al., 2019 and Nielsen et al., 2014.
Other than primary DETCs there is also a 7-17 DETC cell line, which can also be used to study the effect of various factors on DETCs and its interaction with other cutaneous cells. The 7-17 cell line was originally established from FACS purified DETC from AKR mice and expanded by repeated stimulation with concanavalin A (1µg/ml) supplemented with rIL-2 (Nielsen et al., 2014) (Edelbaum, Mohamadzadeh, Bergstresser, Sugamura, & Takashima, 1995)
Preparation of samples for flow cytometer and usage of the machine is a complex process and beyond the scope of this chapter. Before planning the FACS experiment one should be familiar with the general background and theory of flow cytometry (“Practical Flow Cytometry, 4th Edition | Wiley,” n.d.)
Culture of Primary DETCs and 7-17 DETC cell line
(i) Both primary DETCs isolated by FACS and 7-17 DETC stable cell lines are cultured in RPMI 1640 media at 37 °C, 5% CO2. The RPMI medium is supplemented with 10%FBS, 1% L-glutamine, 0.5 IU/L penicillin, 500mg/L streptomycin, 50uM β- mercaptoethanol, 0.63mM HEPES, 1mM Na pyruvate, 1µM nonessential amino acids and 5U/ml IL-2 (Nielsen et al., 2014)(Sharp et al., 2005).
(ii) Primary DETCs and DETC cell lines can be cultured as suspension culture by using ultra-low attachment culture plates. Every two days, remove half of the media by tilting the dish gently allowing the cells to settle down and replenish with fresh media.
(iii) Once cells become 70% confluent, passage them by collecting the total cell suspension into a 15ml tube, centrifuge the cells at 300g for 5 minutes. Remove the supernatant gently and re-suspend the cells in 1ml of fresh media. From this add 200µl to a new 10cm dish containing 10ml of fresh growth media and culture them in the same way as described above.
(iv) In vitro cultures of DETCs are a useful platform to study their interactions with different cell types and the effect of various soluble factors. For example, DETCs can be activated by either cytokines secreted from neighboring cells or by direct cell-cell interactions. It has also been observed that DETCs are activated by co-culturing them with hair follicle stem cells (Badarinath et al., 2019).
While culturing different cell types in co-culturing experiments we should always be careful about the growth conditions of different cell types as inappropriate conditions of culturing any of the cell types can affect the cells in various ways such as stress, proliferation and apoptosis.
(v) We can study the effect of various soluble secreted factors on activation of DETCs. One method utilizes conditioned media from skin explants in which the researchers have examined the effect of IL-1α secreted from wounded keratinocytes on activation of DETCs explained in detail in (Lee et al., 2017)(Badarinath et al., 2019).
For conditioning use the same medium in which the subsequent culture experiments will be performed. Avoid serum in media for these experiments as serum will have its own effect on the cells.
For positive control DETCs are treated with recombinant IL-23(10 ng/ml), recombinant IL-1beta (10 ng/ml), anti-CD3 (1 μg/ml), or combinations of these and scored for activation by looking at the expression of IL-17, FGF7, TNFα and IFN-γ by ELISA or transcript levels as mentioned earlier.
Proliferation assays for activated DETCs in vitro
Elevated proliferation is one of the hallmarks of activated DETCs and can thus be used as a readout for activation. In addition to proliferation other biomarkers of activated DETCs include the expression and secretion of various cytokines such as IL-17, FGF-7, TNFα, and IFN-γ. These secreted cytokines can be detected after 48hrs using an ELISA kit (Nielsen et al., 2014), or are evident at the transcript levels after 24hrs of treatment (Lee et al., 2017).
1. Effect of secreted factors on DETCs
(i) For proliferation assays, culture DETCs as explained earlier. When cells are 70% confluent collect the cells in 15 ml falcon centrifuge at 300g for 5 min.
(ii) Dilute the control and test conditioned media 1:3 with fresh RPMI media.
Preparation of conditioned media is explained in detail in Badarinath et al., 2019.
(iii) Resuspend the cells in the respective conditioned media and plate 30,000 cells in each well of a 96 well dish.
(iv) Incubate the cells in conditioned media for 24 to 48 hours at 37 °C.
(v) After incubation remove the media containing DETCs and quantify proliferation at different time intervals such as 24, 48 and 72
hrs.
There are various assays available by which you can count the number of cells for proliferation assay after treatment with various stimuli such as MTT cell proliferation assay, WST-1 cell proliferation assay, and trypan blue cell counting.
2. Effect of various factors secreted by DETCs on hair follicle stem cells
Isolate primary hair follicle stem cells as previously described in Nowak & Fuchs, 2009
(i) Treat DETCs with conditioned media from control or test skin/epidermal explants animal for 16-24hrs.
(ii) Collect the suspension culture in a 15ml falcon tube and centrifuge at 300g for 5min. Collect the supernatant and discard the cell pellet.
(iii) Dilute the conditioned media 1:5 with fresh E-media.
(iv) Incubate hair follicle stem cells with diluted conditioned media for 24-48 hrs at 37 °C, 7% CO2.
(v) Count the cells at different time intervals over the 24-48 hour time period.
In vivo analysis of DETC activation
The tissue microenvironment strongly dictates the regulation and function of DETCs. Under homeostatic conditions, DETCs have a distinctive dendritic morphology, but after injury or stress, DETCs proximal to the wound site acquire a rounded morphology and transiently lose their dendrites. In addition to morphological changes, another marker of DETC activation is an increased proliferative index. Upon activation, DETCs release certain cytokines that play a significant role in maintaining the protective physical and immune barrier of the murine skin. A variation in DETC function can aggravate skin-related autoimmune diseases, impede tumour eradication, or disrupt proper wound healing (Cruz, Diamond, Russell, & Jameson, 2018). Hence, in vivo analysis of DETCs advances our understanding of the function of these cells in both physiological conditions such as wound healing as well as a variety of pathological scenarios including fibrosis, inflammatory diseases, and carcinomas.
Pipeline to assess DETC activation in vivo:
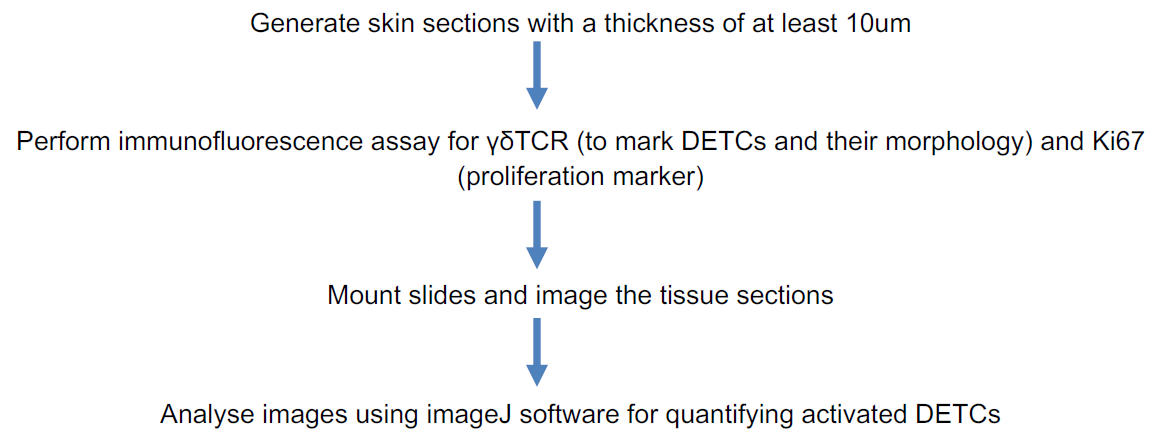
Procedure:
1. Sectioning of mouse skin
(i) To embed skin in OCT medium, follow the protocol described in Gund, Zirmire, J., Kansagara, & Jamora, 2021.
(ii) Section the frozen blocks as described in Fischer, Jacobson, Rose, & Zeller, 2008.
(iii) Store the sections on the charged slides at -80 °C.
Notes:
(a) Collect the skin from the same region of the mice to compare the DETCs between the control and the test animal. It is known that there is heterogeneity of DETCs in different regions of the mouse skin.
(b) Since DETCs are dendritic in morphology when inactive, it is better to have thicker sections to visualize the morphology and quantify dendrites. Hence, take sections of ≥ 10um thickness.
2. Immunofluorescence assay for γδTCR and Ki67
(i) Remove the frozen slides with skin sections from the -80 °C freezer and thaw them at room temperature for at least a minute (but not longer than 5 minutes).
(ii) Using a kimwipe carefully remove the condensation around the skin tissues. Place the slides in a humidifying chamber (Gund, Zirmire, J., Kansagara, & Jamora, 2021).
(iii) Make a hydrophobic barrier around the tissue section using a hydrophobic pen. To fix the sections, add 50ul-100ul of 4% PFA per tissue section for 10 min at room temperature. Make sure the tissue sections are completely covered with PFA.
Notes:
(a) This allows you to minimise the volume of buffers and antibodies being used and gives the ability to differentially stain multiple skin sections on one slide.
(b) Make sure to read the antibody datasheet to utilize the appropriate fixative. This protocol is described for the antibodies mentioned in the reagents section.
(c) Do not exceed the fixation time more than 10min for tissues less than 10um thick as over- fixing leads to excess cross linking of antigens and can produce false negative results.
(d) Thicker sections must be fixed for a longer time - An overnight incubation at 4℃ is recommended.
(iv) Aspirate the 4% PFA after 10 min of incubation. Wash the sections thoroughly with 1X PBS for 5 min each three times.
Note:
a) Aspiration can be avoided if the sections are loosely attached to the slide. Instead, one can remove the fixative/buffer using a pipette.
(v) Add sufficient amount of blocking buffer to cover the tissue sections to block the non- specific interactions for 1 hr at room temperature.
Note:
a) Freeze thawing of the section causes permeabilization of the plasma membrane and the integrity of the membrane proteins is maintained. Hence, additional permeabilization steps with any harsh detergents such as Tween-20 or Triton-X should be avoided as this can disrupt membrane proteins especially if left for too long.
(vi) Incubate the sections with primary antibody (γδTCR to mark DETCs and Ki67 to mark the proliferating cells) diluted in the blocking buffer (refer table 2 for dilutions). Add 50ul - 100ul of the diluted primary antibody on each section and incubate overnight at 4 °C in the humidifying chamber.
Note:
a) Make sure that the hydrophobic barrier is intact. If not, mark the boundaries again with the hydrophobic pen around the sections before adding primary antibody.
(vii) Aspirate the primary antibody and wash the sections with 1X PBS for 5 min each three times.
(viii) Add 50ul-100ul of secondary antibody (refer table 2 for the antibody dilutions) diluted in the blocking buffer onto the sections and incubate for 20 min at room temperature in the humidifying chamber.
Note:
a) γδTCR antibody used in this protocol is a conjugated antibody and does not require a secondary antibody.
(ix) Aspirate the secondary antibody and wash the sections with 1X PBS for 5 min each three times.
(x) Mount the sections with Vectashield mounting media.
Note:
a) Sections can be mounted using alternative mounting media such as Mowiol or 80% glycerol.
(xi) Image the slides under a fluorescent microscope.
Note:
a) Though the signal is optimal if imaged immediately after staining, the stained sections can be stored at 4 °C before imaging the slides.
Alternative method: If using other antibodies along with γδTCR that require additional permeabilization of the tissue, follow the procedure below:
i. For staining of γδTCR, follow the protocol in section B up to step #8.
ii. Repeat steps from #1 to # 5 on the sections.
iii. Add 50ul of permeabilization buffer (0.25% Triton X-100 in PBS) and incubate for 15 min at room temperature.
Note:
a) The concentration of Triton X-100 used for permeabilization should be determined for each antigen of interest.
iv. Block the sections with a blocking solution containing goat serum and 0.1% Triton X 100 for 1 hour at room temperature.
Note:
a) Blocking buffer should contain heat-inactivated normal serum from the same species as the host of the secondary antibody.
v. Incubate the sections with primary antibody diluted in the blocking buffer for 2 hr at room temperature.
vi. Follow steps from # 8 to #12 from section B of the protocol.
Table 2
Summary of various primary and secondary antibodies used in section B along with their respective dilutions.
Sl. No | Source and identifiers | Primary antibody - Dilution | Source andidentifiers | Secondary Antibody - Dilution |
1 | Abcam
AB16667 | Rabbit anti Ki67 – 1:200 | Molecular Probes | Goat antiRabbit Alexa fluor 488 – 1:250 |
2 | Ebiosciences
12-5711-82 | Hamster anti γδTCR PE – 1:100 | - | - |
3 | Jamora lab generated | Chicken anti K5 – 1:200 | Molecular Probes | Goat antichicken Alexa fluor 647 – 1:250 |
4 | - | - | Molecular Probes | DAPI – 1:1000 |
3. Imaging acquisition and analysis:
Image the stained sections under a fluorescent microscope. To quantify the number of proliferating DETCs, lower magnification images of 10x and 20x will suffice. To visualize the morphological differences between the inactive and active DETCs, higher magnification (>40X) images are required.
Note:
a) z-stack images are recommended to fully visualize the dendrites of DETCs that spread throughout the epidermis in multiple planes.
Under homeostatic conditions, DETCs are generally inactive and restricted to the basal layer of the epidermis and the upper region of the hair follicle (Lee et al., 2009). As their name indicates, they possess a dendritic morphology. On the other hand, conditions in which DETCs are activated such as a wound, and mouse models of atopic dermatitis (Lee et al., 2009) (Lee et al., 2017) and squamous cell carcinoma (Du et al., 2010) (De Craene et al., 2014), the cells lose their dendritic extensions and appear more rounded in morphology (Lee et al., 2009) (Du et al., 2010). It has been previously reported that caspase 8 cKO mice, a model for atopic dermatitis (Lee et al., 2009) (Du et al., 2010) exhibits activated DETCs (Lee et al., 2017). In this protocol we demonstrate that this activation of DETCs (both morphological changes and increased proliferation) is conserved across mouse models with a strong inflammatory phenotype. For instance, we have observed that a mouse model for cutaneous squamous cell carcinoma (K14 Snail transgenic mice) (Du et al., 2010), also exhibits activation of DETCs as early as the neonatal stage (Figure 1 and Figure 2).
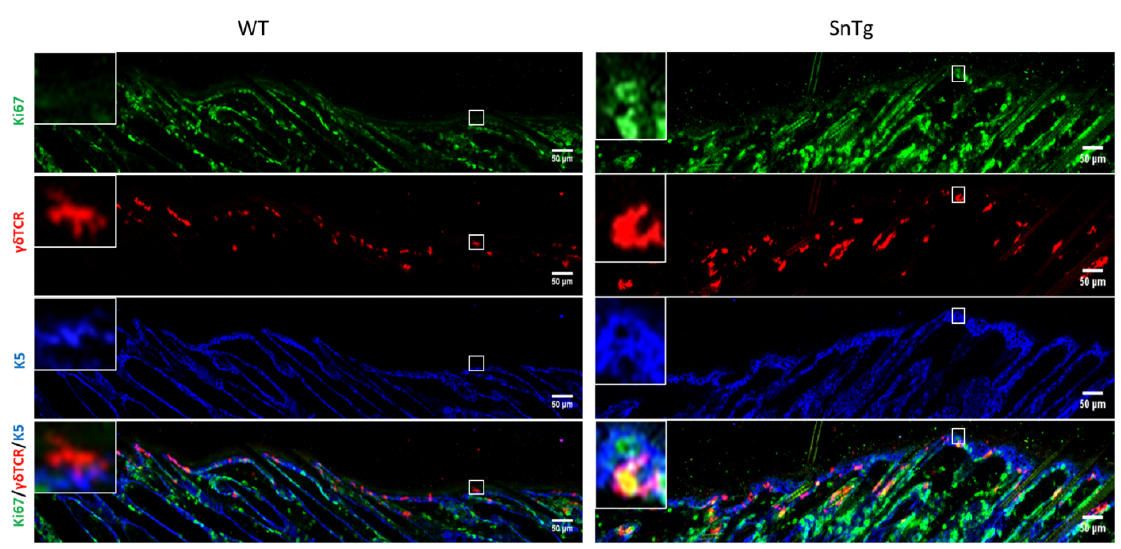
Figure 1. Visualization ofactivated DETCs in WT andSnailTransgenic skin. lmmunofluorescence assay for Ki67 (green) to depictproliferation, γδ15TCR (red) to mark DETCs and K5 (blue) to mark the basal layer ofepidermis on WT and SnTg skin section to observe the activation of DETCs. (Scale bar: 50 um)
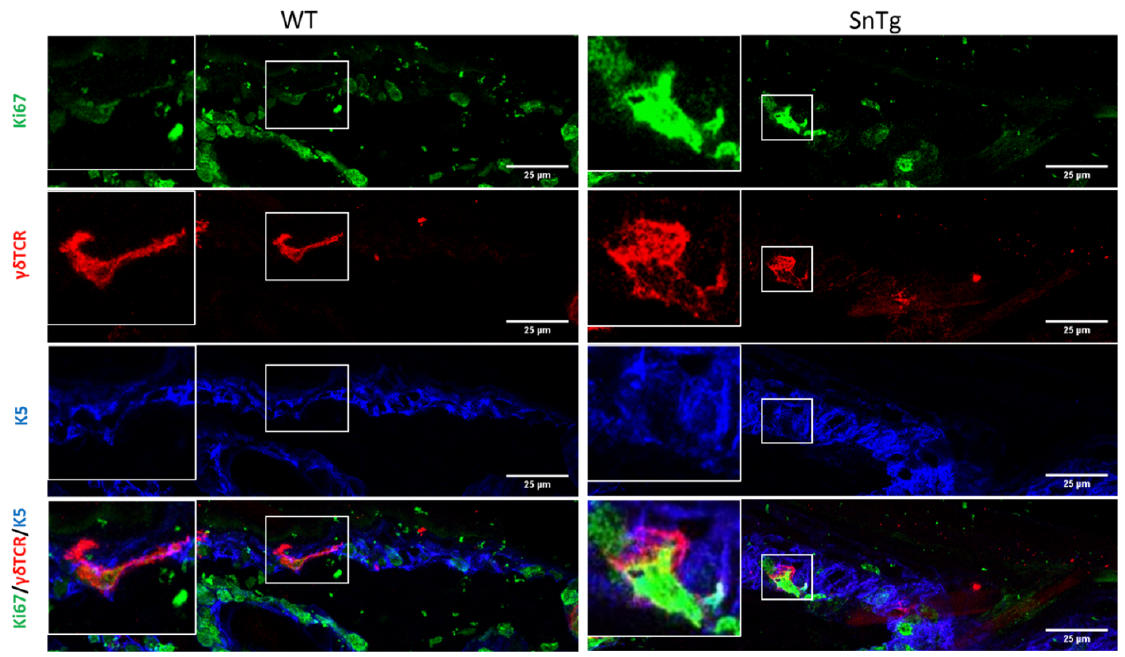
Figure 2. Morphological changes upon activation. Round morphology of DETCs marked by γδTCR (red) in Snail Tg skin shows signs of activation compared to the dendritic morphology of DETCs in WT skin. (Scale bar: 25um)
When activated, DETCs proliferate and hence are Ki67 positive. Therefore, to quantify the number of proliferating DETCs, we counted the number of γδTCR positive cells that are positive for Ki67 across the epidermis of postnatal day 7 - WT and K14 Snail transgenic mice. The number of DETCs across the epidermis of WT and Snail transgenic skin is variable. Hence to ensure consistency in quantification among different backgrounds, we counted all the γδTCR positive cells per 1mm skin section.
Percentage of DETCs that are Ki67 positive and negative were quantified by using ImageJ as follows: Open the merged image of γδTCR and Ki67 on ImageJ software > Main menu > Plugin > Cell counter > Initialize > Type 1. Click on the cells that are positive for both γδTCR and Ki67. This will give the number of DETCs that are proliferating. To count the number of DETCs that are negative
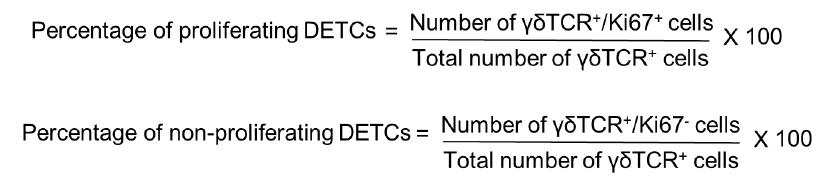
for Ki67 go to ImageJ software>Main menu > Plugin > Cell counter > Initialize > Type 2. Percentage of proliferating and non-proliferating DETCs were calculated using the following formulae:
The proportion of DETCs which are Ki67 positive and negative in WT and K14 Snail Tg skin are as shown in (Figure 3).
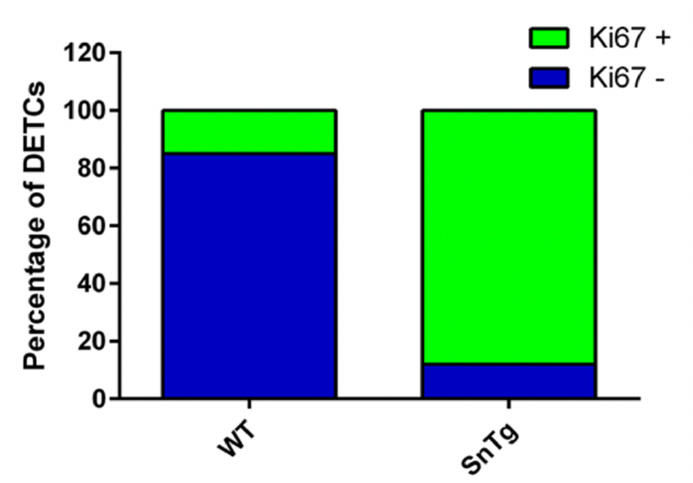
Figure 3. Quantification of activated DETCs in WT and SnTg skin. Percentage plot depicting the proportion of proliferating DETCs which suggests that Snail transgenic skin has more proliferating DETCs compared to WT skin.
To quantify the circularity of the DETCs in WT and SnTg epidermis, ImageJ software was used. Taking the image of just the DETCs (γδTCR positive cells) > Mark the outline using the polygon selection tool on ImageJ for each DETC > Analyze > set measurement > Area and shape description (select “Shape descriptor” parameter to measure and display) > Analyze > Measure or Ctrl+M. This will display a table with measurements. Perform this step for all of DETCs in WT and SnTg skin sections. Circularity value of 1 indicates that a cell is a perfect circle and values that approach 0 indicate that the cell is elongated. Here in this protocol, we have used a threshold circularity value of ≥ 0.5 to call a DETC active. A graph plotted using these measurements (Figure 4) shows that the DETCs in K14 Snail transgenic skin are more circular - indication of activation compared to that in the WT skin.
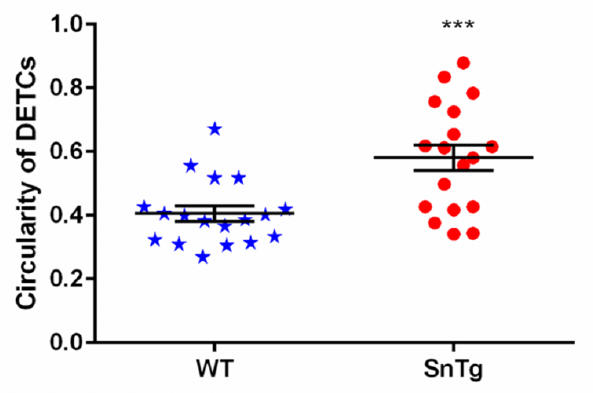
Figure 4. Quantification of morphological changes observed in DETCs. Activated DETCs show morphological changed —from dendritic to circular in shape which was quantified using ImageJ software. Scatter plot shows that DETCs in Snail transgenic epigermis are more circular (circulartity value ≥ 0.5) compared to EDTCs in WT epidermis. Data represent the values ± SEM, ***P < 0.0001, by Student's t-test.
References:
Badarinath, K., Dutta, A., Hegde, A., Pincha, N., Gund, R., & Jamora, C. (2019). Interactions between epidermal keratinocytes, dendritic epidermal T-cells, and hair follicle stem cells. In Methods in Molecular Biology (Vol. 1879). https://doi.org/10.1007/7651_2018_155
Cruz, M. S., Diamond, A., Russell, A., & Jameson, J. M. (2018, June 6). Human αβ and γδ T cells in skin immunity and disease. Frontiers in Immunology, Vol. 9, p. 1. https://doi.org/10.3389/fimmu.2018.01304
De Craene, B., Denecker, G., Vermassen, P., Taminau, J., Mauch, C., Derore, A., … Berx, G. (2014). Epidermal Snail expression drives skin cancer initiation and progression through enhanced cytoprotection, epidermal stem/progenitor cell expansion and enhanced metastatic potential. Cell Death and Differentiation, 21(2), 310–320. https://doi.org/10.1038/cdd.2013.148
Du, F., Nakamura, Y., Tan, T. L., Lee, P., Lee, R., Yu, B., & Jamora, C. (2010). Expression of snail in epidermal keratinocytes promotes cutaneous inflammation and hyperplasia conducive to tumor formation. Cancer Research, 70(24), 10080–10089. https://doi.org/10.1158/0008-5472.CAN-10-0324
Edelbaum, D., Mohamadzadeh, M., Bergstresser, P. R., Sugamura, K., & Takashima, A. (1995). Interleukin (IL)-15 promotes the growth of murine epidermal γδ T cells by a mechanism involving the β- and γ(c)-chains of the IL-2 receptor. Journal of Investigative Dermatology, 105(6), 837–843. https://doi.org/10.1111/1523-1747.ep12326630
Fischer, A. H., Jacobson, K. A., Rose, J., & Zeller, R. (2008). Cryosectioning tissues. Cold Spring Harbor Protocols, 3(8), pdb.prot4991. https://doi.org/10.1101/pdb.prot4991
Gay, D., Kwon, O., Zhang, Z., Spata, M., Plikus, M. V, Holler, P. D., … Cotsarelis, G. (2013). Fgf9 from dermal γδ T cells induces hair follicle neogenesis after wounding. Nature Medicine, 19(7), 916–923. https://doi.org/10.1038/nm.3181
Gund, R., Zirmire, R., J., H., Kansagara, G., & Jamora, C. (2021). Histological and Immunohistochemical Examination of Stem Cell Proliferation and Reepithelialization in the Wounded Skin. BIO-PROTOCOL, 11(2). https://doi.org/10.21769/BioProtoc.3894
Gustafsson, K., Herrmann, T., & Dieli, F. (2020, May 14). Editorial: Understanding Gamma Delta T Cell Multifunctionality - Towards Immunotherapeutic Applications. Frontiers in Immunology, Vol. 11. https://doi.org/10.3389/fimmu.2020.00921
Jameson, J., & Havran, W. L. (2007, February). Skin γδ T-cell functions in homeostasis and wound healing. Immunological Reviews, Vol. 215, pp. 114–122. https://doi.org/10.1111/j.1600-065X.2006.00483.x
Jameson, J., Ugarte, K., Chen, N., Yachi, P., Fuchs, E., Boismenu, R., & Havran, W. L. (2002). A role for skin γδ T cells in wound repair. Science, 296(5568), 747–749. https://doi.org/10.1126/science.1069639
Kashem, S. W., & Kaplan, D. H. (2018). Isolation of Murine Skin Resident and Migratory Dendritic Cells via Enzymatic Digestion. Current Protocols in Immunology, 121(1). https://doi.org/10.1002/cpim.45
Lee, P., Gund, R., Dutta, A., Pincha, N., Rana, I., Ghosh, S., … Jamora, C. (2017). Stimulation of hair follicle stem cell proliferation through an IL-1 dependent activation of γδT-cells. ELife, 6. https://doi.org/10.7554/eLife.28875
Li, F., Adase, C. A., & Zhang, L. J. (2017). Isolation and culture of primary mouse keratinocytes from neonatal and adult mouse skin. Journal of Visualized Experiments, 2017(125). https://doi.org/10.3791/56027
Nielsen, M. M., Lovato, P., MacLeod, A. S., Witherden, D. A., Skov, L., Dyring-Andersen, B., … Bonefeld, C. M. (2014). IL-1β–Dependent Activation of Dendritic Epidermal T
Practical Flow Cytometry, 4th Edition | Wiley. (n.d.). Retrieved February 16, 2021, from https://www.wiley.com/en-in/Practical+Flow+Cytometry%2C+4th+Edition-p-9780471411253 Cells in Contact Hypersensitivity. The Journal of Immunology, 192(7), 2975–2983. https://doi.org/10.4049/jimmunol.1301689
Nowak, J. A., & Fuchs, E. (2009). Isolation and culture of epithelial stem cells. Methods in Molecular Biology, 482, 215–232. https://doi.org/10.1007/978-1-59745-060-7_14
Sharp, L. L., Jameson, J. M., Cauvi, G., & Havran, W. L. (2005). Dendritic epidermal T cells regulate skin homeostasis through local production of insulin-like growth factor 1. Nature Immunology, 6(1), 73–79. https://doi.org/10.1038/ni1152
Witherden, D. A., & Havran, W. L. (2011, January 1). Costimulating epithelial γδ T cells. Cell Cycle, Vol. 10, pp. 4–5. https://doi.org/10.4161/cc.10.1.14290
Related files
 gamma delta activation assay protocol.pdf
gamma delta activation assay protocol.pdf - Rana, I, Badarinath, K, Zirmire, R and Jamora, C(2021). Quantifying γδT-cell activation in vivo and in vitro. Bio-protocol Preprint. bio-protocol.org/prep850.
- Lee, P., Gund, R., Dutta, A., Pincha, N., Rana, I., Ghosh, S., Witherden, D., Kandyba, E., MacLeod, A., Kobielak, K., Havran, W. L. and Jamora, C.(2017). Stimulation of hair follicle stem cell proliferation through an IL-1 dependent activation of γδT-cells. eLife. DOI: 10.7554/eLife.28875
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

