Advanced Search
Purification of mitoribosomes with Oxa1L from HEK cell
Last updated date: Nov 18, 2025 Views: 1204 Forks: 0
Abstract
Mitochondrial ribosomes (mitoribosomes) perform protein synthesis inside mitochondria, the organelles responsible for energy conversion and adenosine triphosphate (ATP) production in eukaryotic cells. To investigate their functions and structures, large-scale purification of intact mitoribosomes from mitochondria-rich animal tissues or HEK cells were developed.
However, the challenge remains, particularly in the fast purification of mitoribosomes anchored to the mitochondrial inner membrane in complex with the Oxa1L translocase. Herein we present a protocol recently developed and modified in our lab, including culturing of HEK293 cells, isolation of mitochondria, and purifications of mitoribosomes with Oxa1L translocase.
Background
Human mitochondria possess their own genome and house all components necessary for transcription, RNA maturation and translation of the encoded genetic information1. Mitoribosome, one of the most important machineries within mitochondria, is responsible for the translation of the essential mitochondrial mRNAs which are coding for components of oxidative phosphorylation (OXPHOS) complexes2. As all of the proteins translated by the human mitoribosome are membrane proteins, understanding the mechanism of co-translational membrane protein insertion is central to understanding mitoribosome translation3,4. In human mitochondria, the oxidase assembly 1-Like (Oxa1L) translocase plays a central role in membrane insertion of mitochondrially encoded products4,5. However, because of unique features of mitochondria, little was known about the molecular and quality control mechanism of mitochondrial translation.
With the structures obtained by single particle electron cryo-microscopy (cryo-EM), opportunities now arise to comprehensively study the molecular mechanisms underlying the translation action and quality control of the human mitoribosome6. Although the existence of quality control in mitochondria was predicted based on analogy with bacteria and eukaryotic cytosols7, biochemical or structural evidences of ribosome-associated quality control in mitochondria remain elusive. We reasoned that any attempt to induce translational stalling might trap mitoribosomes in various stages of the translation cycle in the act of nascent chain insertion into the inner mitochondrial membrane, and generate intermediates suitable for structural analyses by cryo- EM. We therefore purified mitoribosomes from a genetically engineered human cell line that lacks the 2’-5’ phosphodiesterase exonuclease 12 (PDE12). PDE12 facilitates the maturation of mt-tRNA, and therefore a PDE12 knockout can lead to aberrant polyadenylation of the 3’ ends of mt-tRNAs and consequently, mitoribosome stalling8,9. As expected, we observed a substantial proportion of stalled mitoribosomes and discovered a mitoribosome-associated quality control pathway (mtRQC) to rescue the elongational stalling ribosomes10. Our report of the first mitoribosome-associated quality control pathway provides novel insights into regulation of mitochondrial translation. Because the cell needs to detect and respond rapidly even to subtle changes in translation rates, quality control is intimately coupled to elongation. During purification we included GMPPCP, a non-hydrolysable GTP analog, to prevent dissociation of GTPases from mitoribosomes. We have therefore successfully captured 5 additional structures of virtually every elongating ribosomal state. Purifying mitoribosome in complex with its translocase Oxa1L had been challenging since membrane proteins are unstable and tend to dissociated in complicated isolation steps. We added n-Dodecyl β-D-maltoside (β-DDM) along with cardiolipin to solubilize mitoribosomes anchored to the mitochondrial inner membrane via the Oxa1L translocase. We have also trapped the Oxa1L translocase on all our active and stalled ribosomes, thus giving us a first look at co-translational insertion of mitochondrial inner membrane proteins.
This protocol was modified and finalized based on prior methods developed in our lab11,12 and others13–15, as well as our current work10. Herein we provide details for the efficient isolation of intact mitoribosomes with its translocase Oxa1L.We combined cell culture of wilde-type or PDE12(-/-) HEK293S cell lines with high-capacity isolation steps used for the biochemical and structural studies of mitoribosomes and Oxa1L.
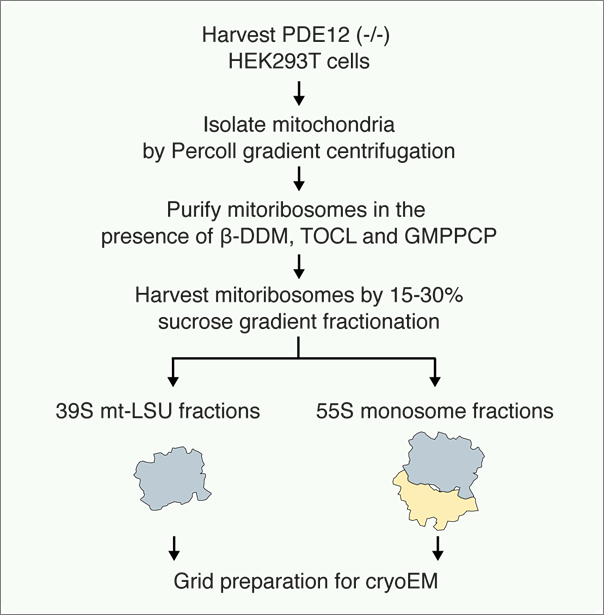
Figure 1. Schematic representations of the procedure
Protocol
1. Cell Culture
1.1 Maintain PDE12-/- HEK cells in DMEM supplemented with 10% FBS, at 37 °C and 5% CO2.
1.2 Scale up to 6 x T175 flasks. At 90% confluency, resuspend the adherent cells in 50-100 ml Freestyle media with 1% FBS; grow in 37 °C, 8% CO2 at 120 rpm.
1.3 Scale up by keeping diluting and splitting with pre-warmed fresh Freesyle+1% FBS media. Count cells using an automated cell counter and adjust cell concentration to 0.5 x 106 cells/mL.
1.4 Count the cells every other day, and proceed to split/dilute the cells if the cell density is above 2.0 x 106 cells/mL.
1.5 Harvest the cells when they reached expected volume with density around 1.0 x 106 cells/mL.
1.6 Harvest cells by centrifugation at 1,000 x g for 7 min, 4 °C. Decant the supernatant carefully and resuspend the pelleted cells quickly in pre-cooled PBS to wash out the media.
1.7 Centrifuge the resuspended cells at 1,200 x g for 10 min, 4 °C. Decant the supernatant carefully and weigh the pellet. Continue to isolate the mitochondria from harvested cells, or snap-freeze the cells in liquid nitrogen and store at -80 °C.
Note: The cell viability should be maintained >90%, and for the final culture that is harvested >95%. The cell culture is specifically for PDE12(-/-) cells. For wild-type cells, all steps are same except two: 1) Grow cells in free style media + 5% FBS in suspension culture (1.3); 2) Harvest cells at concentration ~3-4 x 106 cells/mL. There’re no differences in the following steps. It is important to work quickly and keep everything on ice throughout the procedure.
2. Mitochondria isolation
2.1 Precool the Teflon/glass Dounce homogenizer O/N prior to use.
2.2 Resuspend the pellet (eg 100 g) in 600 mL MIB buffer.
2.3 Allow cells to swell by gently stirring in a 4 °C cold room for 15 min.
2.4 Add 200 mL SM4 buffer (1/3 of MIB buffer).
2.5 Homogenize the samples (>60 up-and-down passes).
(Alternatively, could do nitrogen cavitation with the pressure 500 psi. for 20 min15)
2.6 Centrifuge the homogenized sample at 4 °C, 800 x g, 15min. Collect supernatant (Supernatant-1) through a cheese cloth. Keep the pellet.
2.7 Resuspend the pellet with 200 mL MIBSM (3:1 MIB:SM) buffer, homogenise manually (~15 up-and-down passes); then centrifuge at 4 °C, 800 x g, 15min.
2.8 Collect supernatant (supernantant-2) through a cheese cloth, and combine with Supernatant-1 from step 2.5. Centrifuge at 4 °C, 1,000 x g, 15min. Collect supernatant.
2.9 Centrifuge the supernatant from last step at 4 °C, 10,000 x g, 15min.
2.10 Keep pellet, but carefully wash out loose pellet without disturbing the tight portion.
2.11 Resuspend in 50 ml MIBSM buffer, add 1KU DNase I per 100g cells, leave to rotate on a roller in cold room for 20 min.
2.12 Centrifuge at 4 °C, 10,000 x g, 15min.
2.13 Resuspend the pellet in ~5 mL SEM buffer, homogenize gently with a small dounce homogenizer. Perform no more than five up-and-down passes.
2.14 Prepare 15-60% sucrose gradient in SW28 tubes. Carefully pipetting 4.5 mL of 60% sucrose stock buffer into the bottom of the tube. Carefully add 12 mL of the 32% sucrose stock buffer on top of the 60% band without disturbing it. Repeat with 4.5 mL of 23% sucrose stock buffer and again with 4.5 mL of 15% sucrose stock buffer.
2.15 Load resuspended mitochondrial sample from step 2.13 on sucrose gradient. Centrifuge in SW28 rotor at 4 °C, 28, 000 x rpm, 1h.
2.16 Carefully take the brown band at the interface of 32% and 60% sucrose.
2.17 Snap-freeze the purified mitochondria in liquid nitrogen and store at -80 °C.
Note: Can always start with smaller volume of cells, and subsequent prepare smaller volume of buffers kept the ratio.
3. Mitoribosome purification
3.1 Defrost the frozen mitochondria on ice.
3.2 Add 2 volumes of the lysis buffer to mitochondria (eg. Add 8 mL lysis buffer to 4 mL mitochondria). Mix immediately by inverting the tube several times.
3.3 Homogenize with a small Teflon/glass dounce homogenizer to assist the lysis.
3.4 Leave to rotate on a roller in cold room for 30 min to complete the lysis.
3.5 Centrifuge the lysed material (approximately 9 mL) at 30,000 x g for 20 min, 4 °C to remove the insoluble material. Decant the supernatant carefully from the pellet and discard the pellet.
3.6 Prepare the sucrose cushion in Ti70 (tubes vol approx. 25ml): 7 mL sucrose cushion per tube. Prepare one tube per mL of lysed material.
3.7 Layer lysed mitochondria on the sucrose cushions: approximately 17 mL per tube, resulting in a lysate:cushion ratio of 2.5:1.
3.8 Centrifuge the sample at ~231,550 x g (47k RPM for rotor Ti70) for 60 min at 4 °C.
3.9 Discard the supernatant and rinse the tubes sequentially with 100 µl of the resuspension buffer to remove residual sucrose.
3.10 Resuspend the pellets in total 100 µl resuspension buffer.
3.11 Measure mitoribosome absorption at A260.
3.12 Load the entire sample onto a single linear 15-30% sucrose gradient tube. Centrifuge in TLS-55 rotor at 213, 626 x g (55k RPM for the rotor) for 90 min at 4 °C.
3.13 Fractionate the gradient, determine the optical density at A260 and pool the fraction corresponding to the nucleic acid peak together. The typical A260: A280 ratio of the peak is >1.6.
3.14 Exchange the buffer if necessary, using a method of choice. Calculate the final concentration by using the conversion 1 A260 = 0.1 mg/ mL.
3.15 Use the purified mitoribosome sample for following experiments; Or, snap freeze the purified mitoribosome sample in the resuspension buffer and store at -80 °C.
Materials and Reagents
Dulbecco’s Modified Eagle Medium (DMEM)
tetracycline-free fetal bovine serum (FBS)
Freestyle media (Gibco, Life technologies)
n-Dodecyl β-D-maltoside (D310, Anatrace)
18:1 Cardiolipin (TOCL, 710335C, Avanti)
β,γ-Methyleneguanosine 5′-triphosphate sodium salt (GMPPCP, M3509, sigma)
protease inhibitor (cOmpleteTM, Roche)
rabbit anti-mS27 (Abcam)
mouse anti-OXA1L (Abcam)
Goat anti-mouse 800 (ThermoFisher)
Goat anti-rabbit 680 (ThermoFisher)
MIB Buffer (see Recipes)
SM4 Buffer (see Recipes)
MIBSM Buffer (see Recipes)
SEM Buffer (see Recipes)
60%, 32%, 23%, 15% SG Buffer (see Recipes)
Lysis Buffer (see Recipes)
Sucrose cushion Buffer (see Recipes)
Resuspension Buffer (see Recipes)
15%-30% Linear sucrose gradient Buffer (see Recipes)
Recipes
Buffers for mitochondria isolation
1. Mitochondrial isolation buffer (MIB):
50mM HEPES-KOH, pH 7.4, 10 mM KCl, 1.5mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1mM DTT, protease inhibitors (1 tablet/50 mL).
2. Sucrose/mannitol buffer (SM4):
280 mM sucrose, 840 mM mannitol, 50 mM HEPES-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1m M DTT, protease inhibitors (1 tablet/50 mL).
3. Experimental buffer (MIBSM):
3 volume MIB mix with 1 volume SM4 (e.g. 120 mL MIB buffer + 40 mL SM4 buffer)
4. SEM buffer:
250 mM sucrose, 20 mM HEPES-KOH, pH 7.4, 1 mM EDTA.
5. Sucrose gradient (SG) buffer:
20 mM HEPES-KOH, pH 7.4, 1 mM EDTA and 60% / 32% / 23% and 15% sucrose, respectively. (make stock solutions with 4 different sucrose concentration separately, for preparation of the stepwise sucrose gradient)
Buffers for mitoribosome purification
1. Lysis buffer:
25 mM HEPES-KOH (pH 7.4), 150 mM KCl, 50 mM MgOAc, 1.5% b-DDM, 0.15mg/mL Cardiolipin, 500 mM GMPPCP, 2 M DTT, protease inhibitors (1 tablet/50 mL).
2. Sucrose cushion:
1 M sucrose (34% w/v), 20 mM HEPES-KOH, pH 7.4, 100 mM KCl, 20 mM MgOAc, 0.6% b-DDM, 0.06 mg/mL Cardiolipin, 250 mM GMPPCP, 2 mM DTT.
3. Resuspension buffer:
20 mM HEPES-KOH, pH 7.4, 100 mM KCl, 5 mM MgOAc, 0.15% b-DDM, 0.015 mg/mL Cardiolipin, 250 mM GMPPCP, 2 mM DTT.
4. 15%-30% linear sucrose gradients:
15%-30% linear Sucrose in 20 mM HEPES-KOH, pH 7.4, 100 mM KCl, 5 mM MgOAc, 0.05% b-DDM, 0.005 mg/mL Cardiolipin, 250 mM GMPPCP, 2 mM DTT.
Representative results
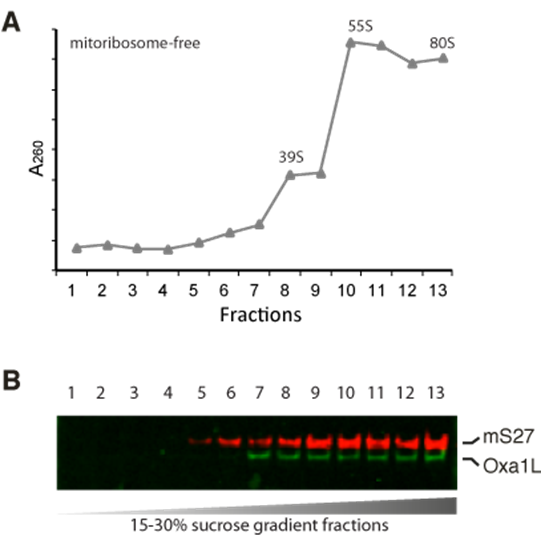
Figure2. purification of mitoribosomes with Oxa1L on a sucrose gradient.
A. Absorbance at 260 nm of the sucrose gradient fractions. B. Western blotting of mitoribosome protein mS27 and OXA1L in fractions.
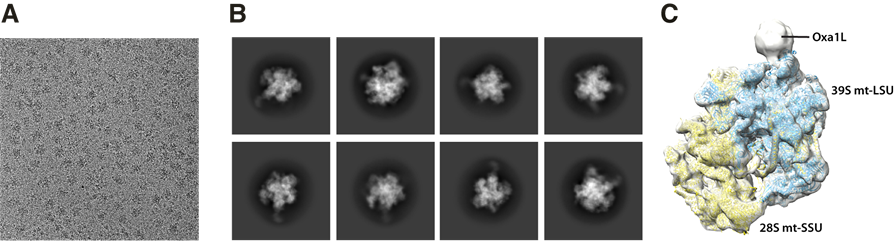
Figure3. cryo-EM micrograph, 2D classes, and 3D initial reconstruction.
A. a micrograph with the mitoribosome-Oxa1L sample. B. A representative post-processing data (2D classes). C. 3D reconstruction.
Acknowledgments
This work received funding from the following sources: UK Medical Research Council (MC_U105184332); Wellcome Trust Senior Investigator award (WT096570); Agouron Institute, and the Louis-Jeantet Foundation; EMBO ALTF 806-2018; Wellcome Trust Clinical PhD Fellowship (110301/Z/15/Z). This protocol was taken from the publication of N. Desai, H. Yang, et al. 2020 with minor modified.
References
1. Dennerlein, S., Wang, C. & Rehling, P. Plasticity of Mitochondrial Translation. Trends Cell Biol 27, 712–721 (2017).
2. Ott, M., Amunts, A. & Brown, A. Organization and Regulation of Mitochondrial Protein Synthesis. Annual Review of Biochemistry 85, 77–101 (2016).
3. Hildenbeutel, M. et al. The membrane insertase oxa1 is required for efficient import of carrier proteins into mitochondria. Journal of Molecular Biology 423, 590–599 (2012).
4. Stiller, S. B. et al. Mitochondrial OXA Translocase Plays a Major Role in Biogenesis of Inner-Membrane Proteins. Cell Metab 23, 901–908 (2016).
5. Haque, M. E. et al. Properties of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. Journal of Biological Chemistry 285, 28353–28362 (2010).
6. Rathore, S. & Ott, M. Structural characterisation of mitochondrial macromolecular complexes using cryo-EM : Mitoribosome biogenesis and respiratory chain supercomplex. (Department of Biochemistry and Biophysics, Stockholm University, 2020).
7. Ayyub, S. A., Gao, F., Lightowlers, R. N. & Chrzanowska-Lightowlers, Z. M. Rescuing stalled mammalian mitoribosomes - what can we learn from bacteria? Journal of cell science 133, (2020).
8. Pearce, S. F. et al. Maturation of selected human mitochondrial tRNAs requires deadenylation. eLife 6, (2017).
9. Rorbach, J., Nicholls, T. J. J. & Minczuk, M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Research 39, 7750–7763 (2011).
10. Desai, N. et al. Elongational stalling activates mitoribosome-associated quality control. Science vol. 370 http://science.sciencemag.org/ (2020).
11. Amunts, A., Brown, A., Toots, J., Scheres, S. H. W. & Ramakrishnan, V. Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015).
12. Brown, A. et al. Structures of the human mitochondrial ribosome in native states of assembly. Nature Structural &Amp; Molecular Biology 24, 866 (2017).
13. Greber, B. J. et al. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 (2015).
14. Greber, B. J. et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 505, 515–519 (2014).
15. Aibara, S., Andréll, J., Singh, V. & Amunts, A. Rapid Isolation of the Mitoribosome from HEK Cells. Journal of visualized experiments : JoVE 1–9 (2018) doi:10.3791/57877.
Related files
 Purification of the Mitoribosome in complex with Oxa1L from HEK Cells.docx
Purification of the Mitoribosome in complex with Oxa1L from HEK Cells.docx - Yang, H and Desai, N(2021). Purification of mitoribosomes with Oxa1L from HEK cell. Bio-protocol Preprint. bio-protocol.org/prep738.
- Desai, N., Yang, H., Chandrasekaran, V., Kazi, R., Minczuk, M. and Ramakrishnan, V.(2020). Elongational stalling activates mitoribosome-associated quality control . Science 370(6520). DOI: 10.1126/science.abc7782
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

