Advanced Search
Splicing efficiency and poly(A) tail length/site
Last updated date: Nov 7, 2020 Views: 956 Forks: 0
Splicing efficiency and poly(A) tail length and site detection.
Day 1. Plate U2OS cells expressing the Doxycycline inducible β-globin reporter in 6 well plates at desired density for transfection the following day.
Day 2. Transfect cells with empty vector or with vector expressing either the WT or S34F mutant U2AF1.
Day 3. 36 hrs after transfection, expression of β-globin reporter was induced for 15 hr. Cells were then scraped and total RNA isolated using the Qiagen RNA isolation kit.
Reverse Transcription and qPCR:
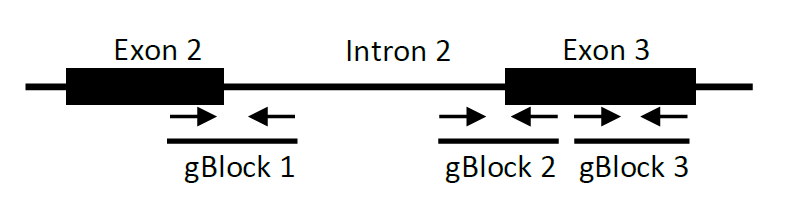
We custom synthesized gene-blocks (double stranded DNA fragments from IDT, Corallville, IA) of the amplicons to be detected in the assay and used them to determine the primer efficiency.
A serial dilution (10-10,000; center the dilution around the concentration that is used in standard PCR reaction) of each gene block was used as template in control qPCR reactions to generate a standard curve for each primer pair. Primer pairs spanning the junction of Exon2-Intron2, and Intron2-Exon3, and a primer pair amplifying a region of Exon3 served to measure unspliced and total RNA of β-globin reporter respectively.
1. 1 μg of total RNA was used to make the first strand using ProtoScript II M-MuLV reverse transcriptase (NEB, Ipsich, MA) and random hexamers (IDT, Coralville, IA) according to the manufacturer's instructions in a final volume of 20 μl.
2. 2 μl of the reverse transcription product was used in a 20 μl qPCR reaction using IQ Syber Green mix (Bio-Rad, Hercules, CA) in a CFX96 qPCR machine (Bio-Rad, Hercules, CA) according to the manufacturer's instructions.
qPCR data analysis:
The unspliced fraction of the reporter RNA was calculated as 10^[(Cqintron-Kintron)/Sintron - (Cqtotal-Ktotal)/Stotal] where Cq is the cycle number, and K and S are the constant and slope respectively of the standard curve for the primer pair. The splicing fractions calculated using Exon2-Intron2 primers or Intron2-Exon3 primers were similar and were therefore combined together. Average
of 4 measurements and associated standard error were calculated.
Poly(A) tail length detection:
Day 1. Plate U2OS cells expressing the Doxycycline inducible β-globin reporter in 6 well plates at desired density for transfection the following day.
Day 2. Transfect cells with empty vector or with vector expressing either the WT or S34F mutant U2AF1.
Day 3. 36 hrs after transfection, expression of β-globin reporter was induced for 15 hr. Cells were then scraped and poly(A)+ RNA isolated using the Qiagen RNA isolation kit.
Poly(A) tail length detection:
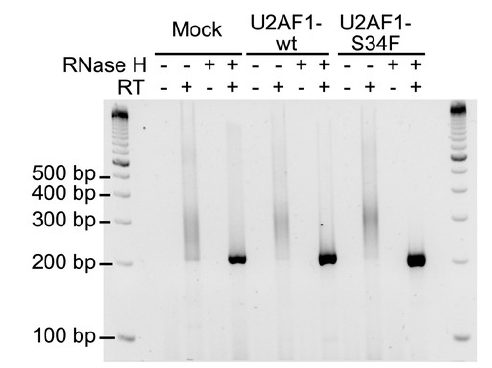
1. 500ng of poly(A)+ RNA was annealed to oligo dT primers (3 fold molar excess of RNA used) and treated with RNAse H (NEB, Ipswich, MA) according to the manufacturer's instructions to remove the poly(A) tails.
2. The digested RNA was then phenol:chloroform extracted, ethanol precipitated and suspended in 10 μl of water.
3. A DNA adaptor (5-GGTCACCTTGATCTGAAGC, with a 5’-phosphate and 3’-amino modification to prevent further ligation; 2 fold molar excess over RNA used) was ligated to 200ng of RNAse H treated RNA (from step 2) or poly(A)+ RNA using T4 RNA ligase 1 (NEB, Ipswich, MA) in a final volume of 20 μl as per the manufacturer’s instructions.
4. 10 μl of the adaptor-ligated RNA was then used to synthesize the first strand using ProtoScript II RT (NEB, Ipswich, MA) and the reverse primer 5ʹ-GCTTCAGATCAAGGTGACC for the poly(A) removed RNA and the primer 5ʹ-GCTTCAGATCAAGGTGACC TTTTT for the poly(A)+ RNA according to the manufacturer's instructions. The reaction products were then digested with RNAse H to remove the RNA.
5. The first stand synthesized (Step 4) was denatured for 20 min at 80°C, and 2.5 μl of the first strand was used as template in a standard PCR reaction using the gene specific forward primer 5ʹ-CCAGGGTTCATCAGATCCTATTCTATAGTGTCAC and the reverse primer 5-GCTTCAGATCAAGGTGACC. Reaction products were then separated on a 3% agarose gel with appropriate DNA size markers to determine the size of the poly(A) tail.
- Larson, D and Palangat, M(2020). Splicing efficiency and poly(A) tail length/site. Bio-protocol Preprint. bio-protocol.org/prep609.
- Coulon, A., Ferguson, M. L., de Turris, V., Palangat, M., Chow, C. C. and Larson, D. R.(2014). Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. DOI: 10.7554/eLife.03939
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
