Advanced Search
Generation of Gadl1-null mice
Last updated date: Nov 3, 2020 Views: 847 Forks: 0
This protocol was performed by Genoway company at Lyon, France.
1)INTRODUCTION
Based on our bioinformatics analysis conditional deletion of the Gadl1 exon 7 (Figure. 1) was chosen as targeting strategy. The strategy results in the conditional deletion of the coding sequences encoding for part of the pyridoxal decarboxylase domain. The splicing of exon 5 to exon 7 will lead to a frame shift resulting in a premature stop codon. The model is generated by homologous recombination in embryonic stem (ES) cells. For this purpose, a targeting vector containing regions homologous to the genomic Gadl1 sequences is constructed.
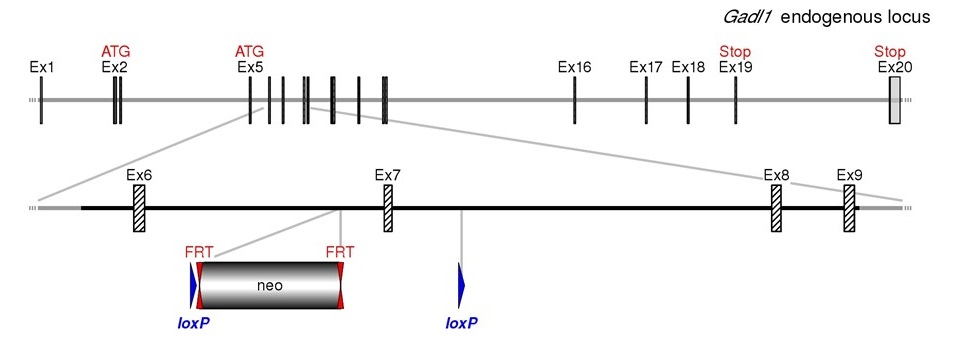
Figure.1. Schematic representation of the selected targeting strategy. Diagram is not depicted to scale.
Hatched rectangles represent Gadl1 coding sequences, grey rectangles indicate non-coding exon portions, solid
lines represent chromosome sequences. The neomycine positive selection cassette is indicated. loxP sites are
represented by blue triangles and FRT sites by double red triangles.
2) GENERAL FEATURES OF THE TARGETING VECTOR
The global strategy for the development of this model is based on the design of a targeting vector
displaying the following features:
Homology regions in C57BL/6 genetic background.
Long homology region of 5.6 kb
Short homology region of 1.5 kb
Insertion of two loxP sites flanking Gadl1 exon 7
Positive selection neomycin cassette flanked by FRT sites. The FRT-flanked selection cassette
can be removed in vivo with recombinase expressing deleter mice
Diphtheria Toxin A (DTA) negative selection marker reduces the isolation of nonhomologous recombined ES cell clones and enhances the chance of isolating ES cell clones harbouring the distal loxP site.
3) ISOLATION OF GADL1 HOMOLOGY REGIONS
This step of the project consists in the cloning of mouse genomic DNA encompassing the murine Gadl1 gene regions surrounding the targeted exons. These isolated sequences are then used to generate
the homology arms necessary for the construction of the targeting vector.
The homologous sequences were cloned from C57BL/6 mouse DNA resulting in the following fragments.
A 4900 bp sized fragment containing exons 8 and 9 and their neighouring intronic sequences.
This fragment is used to generate the distal part of the long homology arm of the targeting
vector. This fragment is referred to as HAA2-LAdi.
A 678 bp sized fragment containing exon 7 and its neighouring intronic sequence. This
fragment is used to generate the proximal part of the long homology arm and is referred to as
HAA2-LApr.
A 1500 bp sized fragment containing exon 7 upstream sequence. This fragment is used to
generate the short homology arm of the targeting vector and to generate a positive control
vector. This fragment is referred to as HAA2-SA.
4)TARGETING VECTOR GENERATION
The targeting vector, and the positive control vector, were successfully generated.
Throughout the construction of the plasmids, each individual cloning step was extensively validated through restriction analysis and partial sequencing.
In addition, the quality of the resulting final targeting vector was controlled using sequencing of:
§ The coding exons
§ The junctions between the homology arms and selection cassette
§ The positive and negative selection cassettes
§ The junction between the homology arm and the plasmid backbone
§ The distal loxP site
5)ESTABLISHMENT OF A PCR SCREENING STRATEGY FOR THE DETECTION OF
THE HOMOLOGOUS RECOMBINATION EVENT
It is absolutely crucial to design screening strategies allowing a quick and unequivocal identification of the homologous recombination event in ES cells.
This screening is performed in two successive steps. First, an initial PCR screening for homologous recombination at the 5' end of the targeting vector is performed. Then, the clones positive for this PCR
based screening are further confirmed by 5’ and 3’ Southern blot analysis.
The initial screening for detection of the expected integration of the targeting vector is achieved by PCR amplification over the 5' short homology arm. This PCR is performed using a primer hybridizing in the neomycin selection cassette (Neo) and a primer hybridizing upstream of the 5' short homology arm in a region absent from the final targeting vector homology sequence. Because of its localisation, this primer set allows unequivocal and specific detection of the 5' integration of the targeting vector in the Gadl1 locus.
For 5' PCR screening set-up, the positive control vector was used as a template. The positive control vector contains the binding sites for both screening primers and mimics the DNA conformation occurring following homologous recombination between the targeting vector and the Gadl1 locus.
6)ESTABLISHMENT OF A SOUTHERN BLOT SCREENING STRATEGY FOR THE
DETECTION OF THE HOMOLOGOUS RECOMBINATION EVENT
As mentioned above, ES cell clones scored positive by PCR screening are further verified by Southern blot analysis. All probes were BLASTed against murine genomic databases in order to select the probes with the best specificity based on in silico analysis. Southern blots were established using wild-type genomic DNA in order to validate probe specificity.
The standard hybridisation conditions used at genOway are indicated below:
Pre-hybridisation and hybridisation: 4 x SSC, 1 % SDS, 0.5 % skimmed milk, 20 mM EDTA,
100 μg/ ml herring sperm, at 65°C for 18 h.
Washings: 2 times 3 x SSC, 1 % SDS at 65 °C for 15 min, then 2 times 0.5 x SSC, 1 % SDS at
65 °C for 15 min.
Exposure: 3 days on BioMax MS films with BioMax intensifying screens.
7)DESIGN OF THE SCREENING STRATEGY FOR THE FLP- AND CRE-MEDIATED EXCISION EVENTS
The Flp-excised allele corresponds to the floxed allele. The Flp-mediated excision enables the deletion of FRT-flanked region. This deletion can be performed in vivo, by breeding the recombined animals with ubiquitous Flp-recombinase expressing deleter mice. Also, the Cre-excised allele corresponds to the Knock-out allele. The Cre-mediated excision enables the deletion of loxP-flanked region. This deletion can be performed in vivo, by breeding the recombined animals with ubiquitous Cre-recombinase expressing deleter mice.
PCR and Southern blot screening was established to enable the wild-type, the recombined, the Flpmediated excised and the Cre-mediated excised alleles to be clearly distinguished.
8)HOMOLOGOUS RECOMBINATION IN EMBRYONIC STEM CELLS
8.1 PREPARATION OF THE TARGETING VECTOR
The targeting vector was linearized by restriction digest with PmeI. The resulting 14.9 kb targeting fragment was isolated and purified by phenol/chloroform extraction followed by ethanol precipitation. This preparation was then used for ES cell electroporation.
8.2 ES CELL TRANSFECTION AND CLONE ISOLATION
The linearised targeting construct was transfected into ES cells according to genOway's standard electroporation procedures (i.e. 5x106 ES cells in presence of 40 μg of linearized plasmid, 260 Volt, 500 μF). Positive selection was started 48 hours after electroporation, by addition of 200μg/ ml of G418. G418 resistant colonies were selected based on cell growth and morphology. 1 electroporation session was performed and a total of 104 clones were isolated and amplified in 96- well plates. Duplicates of 96-well plates were made. One copy was frozen down and stored at -80°C and the other copy of the 96-well plates containing ES cell clones amplified on gelatine was used for genomic DNA preparation and was screened for homologous recombination.
9)SCREENING OF ES CELL CLONES
9.1 PRIMARY PCR SCREENING FOR 5' HOMOLOGOUS RECOMBINATION
Having established the feasibility of the genomic screening, the isolated G418 resistant clones, which were transfected with the targeted vector, were screened using the initial screening PCR to test for homologous recombination at the 5' end of the Gadl1 locus. Recombined clones should yield an amplification product of 1900 bp.
10)RECOMBINANT ES CELL BLASTOCYST INJECTIONS AND GENERATION OF
CHIMERAS
Recipient blastocysts were isolated from pregnant C57BL/6 females (Health status SOPF – specific and Opportunists Pathogens Free). Based on recombined ES cell clones screening results and morphological criteria, recombined ES cell clones were injected into C57BL/6J blastocysts.
Injected blastocysts were then re-implanted into OF1 pseudo-pregnant females (Health status SOPF -
specific and Opportunists Pathogens Free) and allowed to develop to term.
After approximately 3 weeks, offspring were born and the contribution of the recombinant ES cells to
each individual was assessed using coat color markers. Embryonic stem cells are derived from the inner cell mass of 3.5 days old embryos (blastocyst stage). These cells are pluripotent and thus, when implanted into blastocyst-stage embryos, are able to contribute to every cell lineage, including the germ layer. The ES cells used in this experiment were originally derived from a C57BL/6 mouse strain which have black color. These cells were injected into blastocysts derived from an albino C57BL/6 strain (C57BL/6J-Tyrc-2J/J), which have a white coat color. The resulting offspring is thus chimeras of two different cell types (ES cell-derived cells and host blastocyst-derived cells) and the degree of
chimerism can easily be monitored by the percentage of light and dark patches on these animals.
7 highly chimeric males (chimerism rate above 50%) were generated:
11)ASSESSMENT OF GERMLINE TRANSMISSION AND GENERATION OF ANIMALS
11.1 OBJECTIVES
--‐ To investigate whether the recombined ES cell clones have contributed to the germ layer.
--‐ Generation of animals of interest for the project.
11.2 GENERATION OF THE HETEROZYGOUS CONDITIONAL KNOCK-OUT LINE
Breeding were established with C57BL/6 Flp-deleter mice to excise the Neomycin selection cassette and to generate heterozygous mice carrying the Neomycin-excised conditional Knock-out allele.
Progeny was genotyped by PCR. The recombinase mediated excision event was then further validated by Southern blot on a subset of the PCR-positive animals.
11.3 GENERATION OF THE HETEROZYGOUS CONSTITUTIVE KNOCK-OUT LINE
Breeding were established with C57BL/6 Cre deleter mice to excise the loxP flanked region and to
generate heterozygous mice carrying the constitutive Knock-out allele.
- Mahootchi, E(2020). Generation of Gadl1-null mice. Bio-protocol Preprint. bio-protocol.org/prep598.
- Mahootchi, E., Homaei, S. C., Kleppe, R., Winge, I., Hegvik, T., Megias-Perez, R., Totl, C., Mogavero, F., Baumann, A., Glennon, J. C., Miletic, H., Kursula, P. and Haavik, J.(2020). GADL1 is a multifunctional decarboxylase with tissue-specific roles in β-alanine and carnosine production . Science Advances 6(29). DOI: 10.1126/sciadv.abb3713
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
