Thank you for your interest.
RRCS (Residue-residue contact score) can quantitatively evaluates residue contacts based on structural information. The RRCS calculations were done with available python script on https://github.com/elifesciences-publications/RRCS or https://github.com/zhaolabSHT/RRCS
Usage: python RRCS_calculation.py pdb_filename
Output format: Residue_1[Chain ID:Residue ID_Residue] Residue_2[Chain ID:Residue ID_Residue] RRCS
Example: for 2RH1.pdb, run "python RRCS_calculation.py 2RH1.pdb" and got output file "2RH1.pdb.cscore".
The top 10 rows of 2RH1.pdb.cscore is shown below:
A:51_ASN A:79_ASP 2.957555
A:51_ASN A:319_SER 2.449470
A:51_ASN A:80_LEU 0.746105
A:51_ASN A:323_PRO 2.886263
A:51_ASN A:76_ALA 3.645254
A:108_PHE A:170_GLN 0.400661
A:108_PHE A:166_PHE 0.976889
A:108_PHE A:112_ILE 3.025927
A:1054_THR A:1058_ILE 0.580526
A:168_PRO A:199_TYR 1.082637
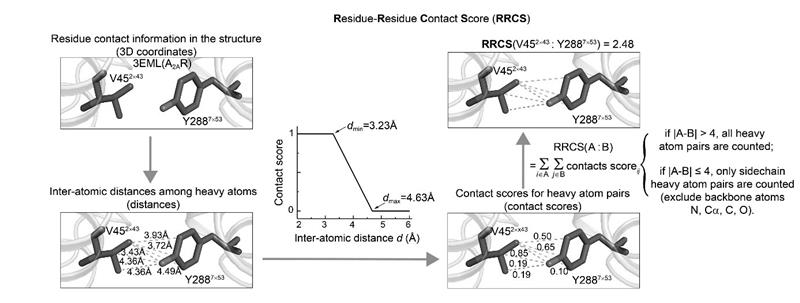
Please see below figure for the workflow of RRCS calculation.
Detailed protocol
1. PDB preprocessing: remove undesired atoms such as fusion proteins, hetero atoms (HETATM) and lipids from the pdb file.
2. Inter-atomic distance calculation: for all possible residue pairs, each possible inter-residue heavy atom pairs are subjected to distance calculation.
3. Inter-atomic contact score calculation: the calculated inter-atomic distances were converted into inter-atomic contact scores based on the score function (shown in above figure).
4. Residue-residue contact score calculation: for the residue pairs between adjacent residues that are within four amino acids in protein sequence, only side chain heavy atom pairs and corresponding inter-atomic contact scores were summated, while atom pairs involving in backbone atoms (Cα, C, O, N) were excluded. For other residue pairs, all possible heavy atom pairs (including backbone atoms) and corresponding inter-atomic contact scores were summated.

