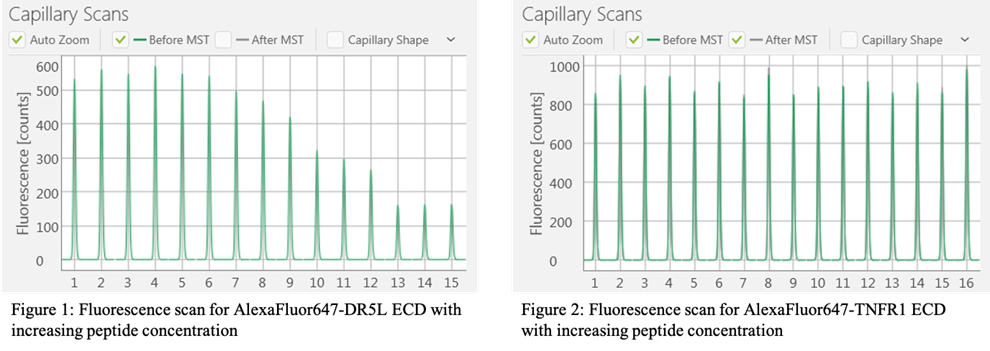
Measuring fluorescence quenching using capillary scanning function of the Monolith NT.115 Instrument
Set-up for this assay was adapted from the MST starting guide by Nanotemper Technologies.
Prepare 2-fold dilutions of 2x unlabeled peptide into 20 mM HEPES pH 7.2, 100 mM KOAc, 0.1% Tween-20, 1% DMSO Distribute 10 ul of 2x unlabeled peptide into PCR tubes Prepare working stock of 400 nM AlexaFluor647-labeled ECD protein in 20 mM HEPES pH 7.2, 100 mM KOAc, 0.1% Tween-20 Add 10 ul of the labeled ECD protein for a final concentration of 200 nM Incubate samples for 30 min at RT in the dark Load samples by capillary action onto Premium capillaries (Nanotemper Technologies, cat. #MO-K025). Scan for fluorescence on a Monolith NT.115 Instrument (NanoTemper Technologies, Germany) at 25°C, medium power.
Important factors to avoid non-specific fluctuations in fluorescence:
Keep % of DMSO constant throughout all samples Use 0.1% Tween-20 in the final sample to prevent aggregation of peptide Make serial dilutions from a singular working stock of peptide Use Premium capillaries (#MO-K025)
Copyright: Content may be subjected to copyright.
How to cite: Readers should cite both the Bio-protocol preprint and the original research article where this protocol was used:
Lam, M(2020). Fluorescence quenching assay. Bio-protocol Preprint. bio-protocol.org/prep397 . Lam, M., Marsters, S. A., Ashkenazi, A. and Walter, P.(2020). Misfolded proteins bind and activate death receptor 5 to trigger apoptosis during unresolved endoplasmic reticulum stress. eLife. DOI: 10.7554/eLife.52291
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this
article to respond.
Post a Question 0 Q&A