Advanced Search
FRET assay
Last updated date: Jan 10, 2025 Views: 317 Forks: 0
Confocal microscopy-based FRET assay with HEK293T cells
Bin Li, Zhenghai Tang, André Veillette*
Abstract
We found that CD2 and its ligands, CD48 in mice and CD58 in humans, were required for T cell activation. These effects occurred independently of CD48 expression on antigen-presenting cells (APCs), implying that CD2 was engaged by ligands present on T cells. Compared to isotype control antibody (Ab), blocking anti-CD2 Ab RM2-5 attenuated the T cell activation responses in the absence of APCs. Here, we describe an assay to determine whether CD2 and CD48 bind in cis, using fluorescence resonance energy transfer (FRET).
Keywords: T cell activation, CD2, CD48, FRET,
Materials and Reagents
HEK293T cells (CRL-3216; American Type Culture Collection, Manassas, VA)
Plasmid vector pFB-Neo (217561; Addgene, Watertown, MA, US) encodes CD2 or CD48, with a SNAP tag or CLIP tag inserted after their amino-terminal signal peptide, respectively.
DMEM (11995-065; Gibco, Waltham, MA, USA)
Fetal bovine serum (12483-020; Gibco, Waltham, MA, USA)
Penicillin-streptomycin-glutamine (10378-016; Gibco, Waltham, MA, USA)
6-well cell culture plates (140675; Thermo Scientific, Waltham, MA, USA)
Accutase (AT-104; Innovative Cell Technologies, San Diego, CA, USA)
Opti-MEM (31985-070, Gibco, Waltham, MA, USA)
Polyethylenimine (PEI) (23966-1; Polysciences, Warrington, PA, USA).
JetPRIME® DNA and/or siRNA transfection reagent (89129-922; VWR International, Mississauga, ON)
Poly-L-lysine (P8920; Sigma-Aldrich, St. Louis, MO, USA)
35 mm dish with glass bottoms (P35G-0-20-C; MatTek Corporation, Ashland, MA, USA)
CLIP-Surface 547 (S9233S; New England Biolabs, Ipswich, MA, USA)
SNAP-Surface Alexa Fluor 647 (S9136S; New England Biolab, Ipswich, MA, USA)
Phosphate buffer saline (PBS)
16% paraformaldehyde (PFA; 15710; Electron Microscopy Sciences, Hatfield, PA, USA)
Purified anti-mouse CD2 Ab (RM2-5; 100102; BioLegend, San Diego, CA, USA)
Purified rat IgG2b, λ isotype ctrl Ab (G013B8; 403801; BioLegend, San Diego, CA, USA)
Equipment and software
An LSM700 confocal microscope, or any confocal microscope equipped with channels for visualizing DY-547, Alexa Fluor 647 and differential interference contrast (DIC). The microscope should support magnification up to 63x for optimal visualization.
Hemocytometer
CO2 incubator
Carl Zeiss Zen, or any other image acquisition software for data acquisition.
ImageJ (Fiji) with the AccPbFRET plugin to analyze the FRET images.
Graphpad Prism was used for statistical analyses.
Procedure
Check the expression of CD2 and CD48 on the surface of HEK293T cells to ensure there is no endogenous expression of these molecules. If endogenous expression is detected, knock out the studied molecules first.
Seed 4x105 HEK293T cells per 35 mm dish (or one well of a 6-well plate) in 2 mL of complete DMEM medium and incubate in a CO2 incubator for 24 hours.
After 24 hours, constructs encoding SNAP-tagged CD2 (0.5 μg) were co-transfected with CLIP-tagged CD48 (0.5 μg) into the cells using the PEI transfection protocol. Scale up the plasmid vector amounts according to the plate format.
Transfection
Prepare DNA molecules and PEI in separate tubes, then transfect HEK293T cells by adding the PEI-DNA mixture to the cells.
Dilute 0.5 μg of SNAP-tagged CD2 and 0.5 μg of CLIP-tagged CD48 plasmids in 100 μL of Opti-MEM and mix gently.
Dilute 3 μg of PEI reagent in 100 μL of Opti-MEM per well and incubate the PEI mixture for 5 minutes at room temperature.
After the 5 minutes incubation, add DNA mix on top of PEI mixture dropwise, tap the solution gently and incubate for 15 minutes at room temperature.
Add the PEI-DNA mixture to the cells dropwise. Mix gently by rocking the plate back and forth.
Incubate cells at 37°C in a 5% CO2 incubator for 8 hours.
After 8 hours of incubation, change the medium with a fresh complete DMEM medium.
After 40-48 hours of transfection, wash the cells with PBS.
Trypsinize the cells with 500 μL of Accutase for 1-2 minutes at 37°C.
Add complete DMEM medium to the cells to neutralize Accutase and make single-cell suspension.
Count and seed 1x105 HEK293T cells onto poly-L-lysine treated 35 mm dishes with glass bottoms.
Incubate the cells at 37°C in a 5% CO2 incubator for 24 hours.
After 24 hours, label cells with CLIP-Surface 547 and SNAP-Surface AF 647. In some cases, a blocking Ab or its isotype-matched control Ab (3 μg/mL) are included.
Cell Surface Labeling Reaction
Dilute the labeling stock solution 1: 200 in medium to yield a labeling medium of 5 μM CLIP-Surface 547 and SNAP-Surface Alexa Fluor 647. Mix dyes thoroughly.
Add 150 μL of the labeling medium to the center of the glass area in each 35mm dish and incubate at 37°C, 5% CO2 for 45 minutes.
Wash the cells three times with 2% FBS in PBS.
Cells are then fixed for 10 minutes at room temperature with 4% PFA and used for the FRET assay.
To remove the PFA, wash the cells three times with PBS.
Images are acquired with an LSM700 confocal microscope by exciting CLIP-Surface 547 (energy donor) at 543 nm and SNAP-Surface AF 647 (energy acceptor) at 635 nm.
Quantification of the FRET efficiency
Install ImageJ and download the AccPbFRET plugin from its original source (Roszik et al., 2008).
Import the two multi-channel images (before bleaching and after bleaching for the same cell) into ImageJ (Fig. 1A).
For each image, split the multi-channel image into individual single-channel stacks. Identify the donor and acceptor images for both the before and after bleaching stages (Fig. 1B).
Subtract average of a background ROI of the images (Fig. 1C).
Generate a FRET image and calculate the value of FRET efficiency according to the steps provided by AccPbFRET plugin (Fig. 1D).
Export the related images and value for analysis (Fig. 2).
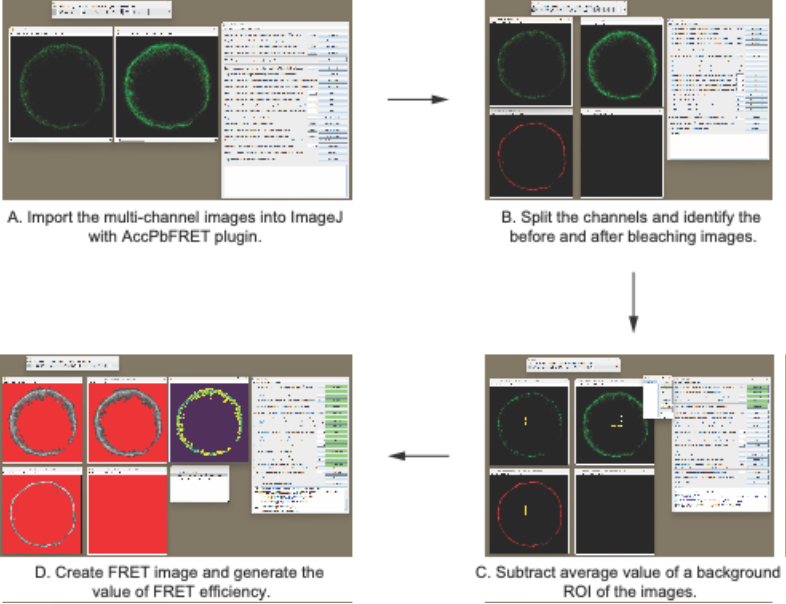
Fig. 1. Stepwise instructions for quantification of FRET efficiency. ROI, region of interest.
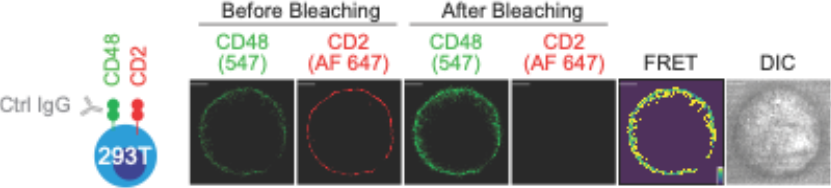
Fig. 2. A FRET assay was used to probe the proximity of CD2 and CD48 by transfecting HEK293T (identified as 293T) cells with CLIP-tagged CD48 and SNAP-tagged CD2. Assay was carried out in the presence of an isotype control IgG. Pseudo-color, the yellow to purple spectrum denotes strong to weak FRET efficiency. Scale bars, 5 μm. DIC, differential interference contrast.
Controls
Negative controls are essential for this analysis and can be implemented using blocking Abs or by generating mutated molecules.
Troubleshooting
Successful calculation of FRET efficiency is influenced by many factors, with the most critical being the preparation of high-quality fluorescence images. In brief, cell images with a clear fluorescent boundaries and minimal background fluorescence are needed. Therefore, ensuring efficient cell transfection is essential. After transfection, FACS can be performed to verify the expression of the studied molecules on HEK293T cells. To troubleshoot transfection efficiency, health and viability of the cell line, quality and quantity of the nucleic acid used, and the choice of the transfection method can all play a part in the outcome of the transfection experiment. Therefore, you may consider using alternative transfection reagents, such as JetPRIME® DNA and/or siRNA transfection reagents, to improve transfection efficiency.
To improve the efficiency of FRET blocking with antibodies, consider adding antibodies during cell seeding onto the glass-bottom plate and incubate overnight.
Copyright: Content may be subjected to copyright.
How to cite:
Readers should cite both the Bio-protocol preprint and the original research article where this protocol was used:
Li B, Tang Z, Veillette A, Confocal microscopy-based FRET assay with HEK293T cells. https://bio-protocol.org/rap50076 (2024).
Li B, Lu Y, Zhong MC, Qian J, Li R, Davidson D, Tang Z, Zhu K, Argenty J, de Peredo AG, Malissen B, Roncagalli R, Veillette A, Cis interactions between CD2 and its ligands on T cells are required for T cell activation. Sci Immunol. 7(74) (2022).
Related files
 Confocal microscopy-based FRET assay with 293T cells (final).docx
Confocal microscopy-based FRET assay with 293T cells (final).docx - Veillette, A, Li, B and Tang, Z(2025). FRET assay. Bio-protocol Preprint. bio-protocol.org/prep2778.
- Li, B., Lu, Y., Zhong, M., Qian, J., Li, R., Davidson, D., Tang, Z., Zhu, K., Argenty, J., de Peredo, A. G., Malissen, B., Roncagalli, R. and Veillette, A.(2022). Cis interactions between CD2 and its ligands on T cells are required for T cell activation. Science Immunology 7(74). DOI: 10.1126/sciimmunol.abn6373
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
