Advanced Search
Taq I Chromosome Conformation Capture (Taq I-3C)
Last updated date: Nov 20, 2024 Views: 181 Forks: 0
Taq I Chromosome Conformation Capture (3C)
Adapted from Chowdhary et al., 2020; Rubio & Gross, 2023; Rubio et al., 2024
Day 1: Overnight Culture
- Inoculate a 10 mL seed culture of S. cerevisiae in an appropriate growth medium. Incubate the culture overnight using a roller drum at 30°C.
Day 2: Treatment and Cell Harvesting
Inoculate the desired volume of fresh growth medium with the overnight culture to an OD600 equivalent of 0.15. Allow cells to grow to mid-log density (OD600=0.65–0.8) with continuous shaking at 30°C. A 50 mL volume of master culture is used per treatment condition, per biological replicate.
NOTE: When comparing strains in the presence or absence of drug treatments, such as Anchor-Away strains in +/− rapamycin conditions, start the treatment at a cell density lower than 0.8. This way cell densities of treated and non-treated strains will be roughly similar at the time of harvesting.
Crosslinking of cells
Perform the desired treatment. Terminate treatment by adding 1.4 mL of 37% formaldehyde (final concentration=1%) to the 50 mL culture. Mix thoroughly with continuous shaking for 15 min at 30°C.
NOTE: If crosslinking is performed at temperatures other than 30°C, appropriate controls must be incorporated to account for any temperature-dependent variation.
- To this, add 8.5 mL of 2.5 M glycine (~375 mM final concentration) to quench excess formaldehyde. Mix thoroughly with continuous shaking for 10 min at 30°C, then place the flask on ice.
- Transfer the cells to a 50 mL tube, and centrifuge at 3000 g for 10 min at 4°C. Repeat pelleting if necessary to collect cells from the whole volume in flask.
- Discard the supernatant and wash the pellet once with 10 mL of ice-cold TBS containing 1% Triton X-100.
- Resuspend the pellet in 1 mL of ice-cold TBS containing 1% Triton X-100 and transfer the cell suspension to a 1.5 mL microfuge tube.
- Pellet cells by centrifugation at 10,000 g for 10 min at 4°C. Discard the supernatant.
- Resuspend the pellet in 500 µL of FA lysis buffer by pipetting up and down. Do not vortex. Add PMSF (Final conc.: 1 mM) to the FA lysis buffer just before use.
Store cells at −80°C or proceed to cell lysis.
Day 3 (2-3 h): Cell lysis and isolation of cross-linked chromatin
Thaw the cell pellet and add an equal volume (600 µL: Cell pellet has a volume ~100 µL) of acid washed, cold (4°C) glass beads. Vortex the cells using 4 cycles of 10 min vortexing, 5 min ice break, at 4°C (full speed setting in Eppendorf MixMate: 3,000 rpm – keep at 4°C the entire time). Leave cells in ice break for 10 min after the second cycle. Flip tube to mix contents before returning to vortexer.
NOTE: Use ice-cold glass beads to limit changes that could arise from heat generated upon vortexing.
- After vortexing, flip the sample tube upside down, clean the bottom of the tube with 70% ethanol and then pierce its bottom using a new 23-gauge needle/sample. Make sure there is nothing close to the bottom of the tube before piercing it (i.e., all contents should be settled underneath cap).
Put the pierced tube upright on top of a new chilled 1.5 mL microfuge tube.
NOTE: Put the puncture hole at the top outer side of the tube when inserted into the rotor: cell lysate will flow easier to receiving tube.
- Spin down the nested microfuge tubes at 4°C for ~5-10 sec. Cell extract will be transferred to the bottom tube; only glass beads will remain in the tube at the top. Resuspend cell lysate by pipetting.
Centrifuge extract at 13,000 g for 10 min at 4°C.
NOTE: Chromatin will be visible as a thin, transparent, glassy layer on top of cell debris.
- Carefully aspirate and discard the supernatant, resuspend the pellet in 1 mL of FA lysis buffer + PMSF (maintain at 0-4°C). Centrifuge again at 13,000 g for 10 min at 4°C. Remove supernatant.
Resuspend the pellet in 500 µL of cold 10 mM Tris-HCl (pH 7.5).
NOTE: Pellet has jelly-like consistency, resuspended slowly by using pipette tip, not by vortexing. Avoid spilling the samples.
Aliquot samples into cold 5 mL tubes, as follows:
- 3C (Ligated) Sample (LIG): 200 µL
- No Ligation (Digested Only) Control (DO): 200 µL
- Undigested Control (UND): 100 µL
NOTE: There is ~100 µL leftover.
- Store leftover cell lysate and aliquots at -80°C.
Day 4 (12 h): Restriction digestion of crosslinked chromatin
- Thaw tubes on ice. To each aliquot of DO and LIG chromatin sample, add 40 µL of 10X restriction enzyme buffer (CutSmart Buffer) and 120 µL of ddH2O. Mix well by pipetting up and down.
Add 800 U (40 μL) of the restriction enzyme Taq I (NEB, Cat # R0149L), mix gently (NO VORTEXING) and incubate at 60°C for 7 hrs. Flip tube every hr to prevent settling.
NOTE: Time and temperature conditions specified in the protocol were optimized for Taq I Restriction Enzyme.
Alternatively, digestion can be performed overnight (16 hrs) after cell lysis. If O/N digestion is done, use device with shaking capabilities. We use Eppendorf ThermoMixer C with the following program: 1 hr OFF, 1 min ON, 700 rpm, for 16 hrs at 60°C.
Terminate digestion by adding 40 µL of 10% SDS (1% final conc., mix well) and incubate the sample at 80°C for 20 min. Then add 2260 µL of ddH2O and 300 µL of 10% Triton X-100 to remove SDS. Mix well.
NOTE: Ligation is inhibited by the presence of SDS. Triton X-100 quenches SDS.
Centrifuge the sample at 13,000 g for 20 min at room temperature (avoid low temp., which precipitates SDS). Discard the supernatant and resuspend the pellet in 400 µL of 10 mM Tris-HCl (pH 7.5).
NOTE: Pellet is still jelly-like in consistency, which allows vacuum aspiration of the supernatant without loss of pellet.
Intramolecular ligation of crosslinked chromatin:
- To LIG tubes, add 1400 µL of Quick ligase buffer (Quick Ligation Kit, NEB, Cat # M2200L; see recipes), 980 µL of ddH2O and 20 µL of the Quick T4 DNA ligase enzyme to 400 µL of the restriction digested chromatin. Mix all the components by pipetting and incubate at 25°C for 2 hr.
- To DO tubes, add 2400 µL of ddH2O and proceed to reversal of chromatin crosslinks.
- Thaw UND tubes on ice, add 1300 µL of ddH2O, proceed to reversal of chromatin crosslinks.
Reversal of chromatin cross-links:
- Add 8 µL of 10 mg/mL DNase-free RNase (30 ng/µL) to DO and LIG tubes (add 4 µL to UND tube) and incubate for 40 min at 37°C.
- Add 28 µL of 10% SDS (final conc.= 0.1%) and 20 µL of 10 mg/mL Proteinase K (final conc.= 70 ng/µL) to the DO and LIG tubes (add 14 µL of SDS and 10 µL of Proteinase K to UND tubes), and incubate O/N (16 hrs, 12-14 hrs minimum) at 65°C.
Day 5 (8 hr): Precipitation of deproteinized genomic DNA
- Transfer LIG and DO samples to 15 mL tubes.
- Add equal volume (for LIG and DO add 2.8 mL; for UND add 1.4 mL) of Phenol:Chloroform:Isoamyl Alcohol (25:24:1) (organic phase 1) to the sample. Vortex for 2 min (Vortex-Genie2, max speed) and then centrifuge at 10,000 g for 10 min at room temperature. Use the appropriate tube adaptors for centrifugation in a Fiberlite F15-8x50c rotor (Thermo Fisher Sci.) or equivalent. Centrifuge in Sorvall Legend X1R.
- Collect the aqueous phase and transfer it to the appropriate tube size. Repeat previous step (organic phase 2).
- Add Chloroform:Isoamyl Alcohol (24:1) to each sample, equal volume, and centrifuge at 10,000 g for 10 min at room temp. (organic phase 3).
- Transfer aqueous phase to a new and clean 15 mL tube.
- To maximize the yield of deproteinized DNA, back extract by adding 1.4 mL of 1X TE to DO and LIG (700 μL to UND) in the leftover interface and organic phase 1. Mix thoroughly as before and centrifuge at 10,000 g for 10 min. Transfer to the next organic phase and repeat until completing back extraction from all organic phases. Collect the aqueous phase and add it to the previously obtained aqueous phase from the same tube.
- To the pooled aqueous phase, add 1/10th volume of 3M sodium acetate (pH 5.2) and glycogen (20 mg/mL): 8 µL of glycogen for DO and LIG, 4 µL for UND. Vortex briefly. Now, add 2.5 vol of ethanol and aliquot 5 mL from each sample into 5 mL tubes. Incubate at room temp. for 40 min (recommended incubation time: 40 to 60 min). Centrifuge for 20 min at 13,000 g at room temp. Discard the supernatant. Repeat aliquoting, centrifugation, and discarding of supernatant until all volume for each sample has been pelleted into the 5 mL tube.
- Centrifuge again for 1 min at 13,000 g at RT. Aspirate the remaining supernatant and air dry the pellet, do not over dry.
- Dissolve the pellet in 500 µL of 1X TE (pH 8.0) for LIG and DO, 250 µL for UND. Store the sample at 4°C for several months.
- Analyze samples by qPCR.
Locus-specific PCR
Perform the locus-specific RT-qPCR reaction using a specific tandem primer pair combination.
To the 96 well qPCR plate, add per well:
10 μL of 2X SYBR Green Master Mix.
0.5 μL each primer
2 μL of the test sample
7 μL of ddH2O
(Adjust volume to the desired number of samples.)
- Obtain the Ct values or the absolute amount of the amplicon generated and normalize it to the amount of amplicon generated using a convergent primer pair mapped to a genomic sequence that lacks a restriction site. Determine the relative frequency of interaction of each primer pair used in the 3C analysis.
Controls
No Template Control
qPCR without 3C template. Allows removal of noise from the formation of primer dimers.
No Ligation/Digested Only Control (DO)
All steps performed except for ligation.
Recovery Control
Use convergent primers used for standard curve using genomic DNA. Recommended target sequence: ARS504, which lacks a Taq I restriction site. Allows for comparison of 3C signals between different experiments.
Digestion Efficiency Control
Efficiency of digestion is calculated for each restriction site evaluated in the 3C analysis. Convergent primers are used. Both undigested (UND) and digested only (DO) templates are analyzed for each primer combination and the Ct values obtained are placed into the following formula:
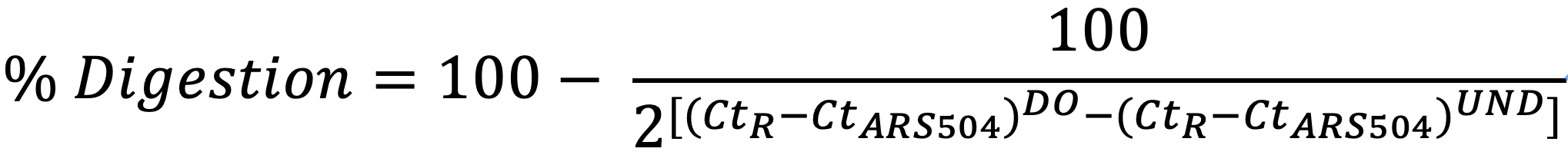
CtR= Cycle threshold quantification of DO or UND templates
CtARS504= Ct value of non-cleaved ARS504 locus for DO or UND templates
Primer-Pair Efficiency Control
Ratio of ligation-dependent signal for 3C over ligation-dependent signal for control template gDNA.
To calculate the frequency of interaction between two chromosomal loci, use the algorithm previously described (Chowdhary et al., 2020) with one modification, namely, that the DO signal is subtracted from the LIG signal rather than generating a LIG/DO quotient (step 6).
Specifically:
Step by step quantification
| Description | Terms and equation |
| 1. Average cycle threshold (Ct) | a. Avg. Ct for DO3C template b. Avg. Ct for Lig3C template =
|
2. Net Ct values This control corrects for background noise due to primer dimers.
| a. Δ b. Δ |
3. Average fold signal above background
| a. Avg. fold signal for DO3C template = b. a. Avg. fold signal for Lig3C template = |
4. Amount of PCR product from ARS504 locus
| a. ARS504(in ng) for DO3C template = b. ARS504(in ng) for Lig3C template = |
5. Fold-over signals normalized to internal control locus This control corrects for the variation in recovery of templates.
| a. Fold over normalized signal for DO3C template = b. Fold over normalized signal for Lig3C template = |
6. Ligation-dependent 3C signal
| ( |
7. % digestion of each participating Taq I site
| % Digestion of Site 1 or 2 = |
8. Ligation-dependent 3C signals corrected for variation in Taq I digestion efficiencies This control corrects for variation in Taq I digestion efficiencies.
| |
9. Corresponding correction for the gDNA template (see gDNA prep protocol below) Note: In our hands, digestion of gDNA is almost always close to 100%.
| |
10. Normalized frequency of interaction The control corrects for variation in primer pair efficiencies as well as the inherent tendency of any two DNA ends to be ligated.
| 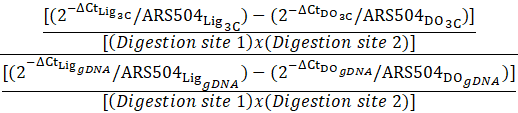 |
Taq I Chromosome Conformation Capture Buffers
5X TBS
50 mM Tris-HCl
1 M NaCl
Set pH to 7.5 using HCl.
FA lysis buffer
50 mM HEPES.KOH (pH 7.9)
140 mM NaCl
1 mM Na-EDTA (pH 8.0)
1% Triton X-100
0.1% Sodium deoxycholate
1 mM PMSF (always add fresh before use)
1X TE buffer
10 mM Tris-HCl (pH 8.0)
1 mM Na-EDTA (pH 8.0)
Other solutions (stock preparations)
2.5 M Glycine
3 M Sodium acetate
1 M Tris-HCl (pH 7.5)
10% SDS
10% Triton X-100
10 mg/mL Proteinase K
10 mg/mL RNase
100 mM PMSF
2x NEB Ligation Buffer*
132 mM Tris (pH 7.6)
20 mM MgCl2
2 mM DTT
2 mM ATP
15% PEG (MW 6000)
Filter sterilize.
*Prepare individual stock solutions for DTT, ATP, Tris-HCl, and MgCl2.
1 M Dithiothreitol (DTT)
Dissolve 1.5 g of DTT (DL-dithiothreitol, anhydrous m.w.=154.25) in 8 mL of deionized or distilled H2O. Adjust volume to 10 mL, dispense into 1-mL aliquots, and store in the dark (wrapped in aluminum foil) at –20°C (indefinitely).
NOTE: Do not autoclave DTT or solutions containing it.
100 mM ATP
1 g ATP (adenosine triphosphate)
12 mL H2O
Adjust pH to 7.0 with 4 M NaOH
Adjust volume to 16.7 mL with H2O
Store in aliquots indefinitely at −20°C. Do not autoclave, Filter sterilize final solution.
> I added the solution of ATP, 0.1 M, without adjusting pH. Final pH of ligase buffer was still neutral. Check pH again.
PEG
Powder can be added to the buffer at the moment of mixing solutions.
Generation of the Naked DNA Control Template ("Dekker Control")
Adapted from Singh et al., 2009.
Purpose: To generate a naked genomic DNA (gDNA) control template for normalization of 3C values. Permits a quantitative comparison of interaction frequencies derived from chromatin templates.
NOTES:
- Use two biological replicates.
- Amount of DNA needed per biological replicate: ≥250 µg
- 10 tubes, with 10 µg each are going to be used for Digestion (DO)
- 10 tubes, with 10 µg each are going to be used for Digestion + Ligation (LIG)
- At least 50 µg of DNA for each biological replicate should be left undigested to determine percentage of digestion (UND)
- Here it is assumed that each restriction site is 100% digested; therefore, a determination of % DIG is not necessary.
Day 1: Primary Culture
- Grow 300 mL culture of the yeast strain O/N at 30°C.
- Grow culture from a large colony, allow growth for at least 18 hrs.
For slowly growing strains, a 10 mL O/N starting culture can be used to seed 300 mL of growth medium.
Day 2: gDNA extraction
Centrifuge culture for 5 min at 3500 g at room temp. Wash the pellet with 10 mL of sterile ddH2O. Resuspend the pellet in 1 mL of ddH2O. Distribute the cell suspension into 1.5 mL microfuge tubes, 100 µL/tube. Obtain, at least, 60 tubes.
Note: 100 mL culture yields ~20 tubes
- To each of the 1.5 mL microfuge tubes containing 100 µL of culture, add 300 µL of ice-cold glass beads.
- Add 200 µL of cell-lysis buffer.
- Add 200 µL of Phenol:Chloroform:Isoamyl alcohol (25:24:1) saturated with Tris-HCl (pH=8.0).Vortex for 10 min at 4°C.
- Add 300 µL of cell lysis buffer and 300 µL of Phenol:Chloroform:Isoamyl Alcohol (25:24:1) solution. Centrifuge at 10,000 g for 10 min at RT.
- Take the aqueous phase and keep repeating the Phenol:Chloroform:Isoamyl Alcohol (25:24:1) extraction until you get a clear aqueous phase.
- Most of the time a single extraction is enough.
- Pool in the aqueous phase from all the microfuge tubes into a 50 mL tube and add 0.6 vol of isopropanol.
- Invert the 50 mL tube several times and incubate for 30 min at RT.
- Centrifuge the sample at 3500 g for 10 min at room temp. Discard the supernatant and air dry the pellet.
- Dissolve the pellet in 1.5 mL of 1X TE (pH=8.0) and transfer it to a 5 mL tube.
- Add 15 µL of DNase-free RNase (10 mg/mL) and incubate for 30 min at 37°C.
- Add 15 µL of Proteinase K (10 mg/mL), mix the tubes gently and leave for incubation at 65°C for 2 hrs.
- Add equal volume (1500 µL) of Phenol:Chloroform:Isoamyl Alcohol (25:24:1) to the reaction mix, invert the tubes several times, and centrifuge at 10,000 g for 10 min. Collect the aqueous phase in a fresh 5 mL tube.
- In order to maximize the yield of deproteinized gDNA, ‘back extract’ by adding 300 µL of 1X TE to the leftover interface and organic phase. Mix thoroughly and centrifuge at 10,000 g for 10 min. Collect the aqueous phase and add to the previously obtained aqueous phase from the same tube.
- Pool the aqueous phase together and add equal volume of isopropanol to it. Keep at room temp. for 30 min.
- Centrifuge at 10,000 g for 10 min at room temp. Discard the supernatant, centrifuge again at 10,000 g for 10 min at room temp. Remove residual isopropanol with pipette tip and leave the pellet for air drying.
- Dissolve the pellet in 1.5 mL of 1X TE (pH = 8.0).
- Record the DNA concentration.
Day 3: Restriction digestion
- Prepare 10 microfuge tubes containing each one 10 µg of genomic DNA. Add 20 µL of 10X restriction enzyme buffer, 50 U of the same restriction enzyme (5 U/µg of gDNA) as used for 3C analysis (Taq I). Bring the final volume in each tube to 200 µL by adding sterile ddH2O. Incubate the tubes at 65ºC for 5 hr.
- Per biological replicate
- DO: 10 tubes, 10 µg/tube
- LIG: 10 tubes, 10 µg/tube
- Per biological replicate
- Pool the contents of each of the ten tubes containing digested chromatin, 2 mL total, into a 5 mL tube. Add equal volume of Phenol:Chloroform:Isoamyl Alcohol (25:24:1). Invert the tubes several times and centrifuge at 10,000 g for 10 min at RT. Collect the aqueous phase and transfer it to a 15 mL tube.
- Perform back extraction by adding 500 µL of 1X TE to the leftover interface and organic phase. Mix thoroughly (vortex for 2 min) and centrifuge at 10,000 g for 10 min. Collect the aqueous phase and add it to the previously obtained aqueous phase from the same tube.
- To the pooled aqueous phase, add 1/10th volume of 3M sodium acetate (pH=5.2) and 2.5 vol of ice-cold ethanol. Incubate for 30 min in ice.
- Centrifuge the samples at 10,000 g for 20 min at 4°C. Discard the supernatant. Wash the pellet by adding 1 mL of 70% EtOH gently and flip over tubes several times gently to wash well.
- Centrifuge again at 10,000 g for 15 min at 4°C. Discard the supernatant, re-centrifuge again at 3500 g for 1 min at 4°C. Remove residual EtOH with pipette tip and leave the pellet for air drying.
- DO: Dissolve the pellet in 200 µL of TE (pH=8.0), transfer to 2 mL tube. Store until LIG samples are completed.
Resuspend the pellet in 45 µL of ddH2O (or Tris-HCL pH=8.0 if DNA is going to be ligated the next day), transfer soln. to a 2 mL tube.
- Can store DNA for next day at this point.
Ligation and Final DNA Extraction
- Add 50 µL of 2X Quick T4 ligase buffer and 5 µL of Quick T4 ligase enzyme (single tube per biological replicate). Mix by pipetting and incubate the sample for 2 hrs at 25°C.
- Add 100 µL of ddH2O to it after incubation.
- Add equal volume (200 µL) of Phenol:Chloroform:Isoamyl alcohol (25:24:1) to the sample, mix it and centrifuge at 10,000 g for 10 min at RT. Collect the aqueous phase.
- Perform back extraction by adding 200 µL of TE to the leftover interface and organic phase. Mix thoroughly and centrifuge at 10,000 g for 10 min. Collect the aqueous phase and add to the previously obtained aqueous phase from the same tube.
- To the pooled aqueous phase, add 1/10th volume of 3M sodium acetate (pH=5.2). Add 2.5 vol of ice-cold ethanol and incubate for 30 min in ice.
- Centrifuge at 13,000 g for 20 min at 4°C. Discard the supernatant, centrifuge at 13,000 g for 1 min at 4°C, remove supernatant. Wash the pellet once with 70% ethanol by adding 1 mL of 70% EtOH gently and flip over tubes several times gently to wash well.
- Centrifuge the sample at 10,000 g for 15 min at 4°C. Discard the supernatant, centrifuge again at 10,000 g for 1 min at 4°C. Remove residual EtOH with pipette tip and leave the pellet for air drying.
- Dissolve the pellet in 200 µL of TE (pH=8.0).
- Store the Dekker’s Control template at -80°C (or -20°C).
Dekker Control Buffers
Cell lysis buffer
10 mM Tris-HCl (pH 8.0)
100 mM NaCl
10 mM Na-EDTA (pH 8.0)
1% SDS
1X TE buffer
10 mM Tris-HCl (pH 8.0)
1 mM Na-EDTA (pH 8.0)
2x NEB Ligation Buffer
132 mM Tris (pH 7.6)
20 mM MgCl2
2 mM DTT
2 mM ATP
15% PEG (MW 6000)
Other solutions (stock preparations)
3 M Sodium acetate
10 mM Tris-HCl (pH 8)
References
Chowdhary, S., Kainth, A. S., & Gross, D. S. (2020). Chromosome conformation capture that detects novel cis- and trans-interactions in budding yeast. Methods, 170, 4–16. https://doi.org/10.1016/j.ymeth.2019.06.023
Rubio, L. S., & Gross, D. S. (2023). Dynamic coalescence of yeast Heat Shock Protein genes bypasses the requirement for actin. Genetics, 223(4). https://doi.org/10.1093/genetics/iyad006
Rubio, L. S., Mohajan, S., & Gross, D. S. (2024). Heat Shock Factor 1 forms condensates and restructures the yeast genome before activating target genes. ELife, 12, RP92464. https://elifesciences.org/reviewed-preprints/92464
Singh, B. N., Ansari, A., & Hampsey, M. (2009). Detection of gene loops by 3C in yeast. Methods, 48(4), 361–367. https://doi.org/10.1016/j.ymeth.2009.02.018
Related files
 Taq I-3C LR Modif. Nov-20-24.docx
Taq I-3C LR Modif. Nov-20-24.docx - Rubio, L S and Gross, D S(2024). Taq I Chromosome Conformation Capture (Taq I-3C). Bio-protocol Preprint. bio-protocol.org/prep2762.
- Rubio, L. S., Mohajan, S. and Gross, D. S.(2024). Heat Shock Factor 1 forms nuclear condensates and restructures the yeast genome before activating target genes. eLife. DOI: 10.7554/eLife.92464
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
