Advanced Search
Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases
Last updated date: Oct 17, 2024 Views: 170 Forks: 0
1.Fungal RNA Extraction Using Modified Trizol Method
Reagents:
a.Trizol
b.Chloroform
c.Isoamyl alcohol
d.Isopropanol
e.70% Ethanol (prepared with anhydrous ethanol and RNase-free water)
f.RNase-free water
Equipment:
a.2 mL EP tubes
b.1.5 mL EP tubes
c.Homogenizer
d.Centrifuge
e.Laminar flow hood
f.UV-Vis spectrophotometer for measuring OD260/280
Fungal RNA Extraction:
1.1 Sterilize a steel bead by high temperature and pressure, then add it to a 2 mL centrifuge tube free of RNase. Add approximately 0.3 g of mycelium (for mycelium obtained from liquid culture, use vacuum filtration; for mycelium from solid culture medium, scrape with a pipette tip), and add 1 mL of Trizol (the amount of mycelium should not exceed 10% of the Trizol volume to avoid DNA contamination).
1.2 Homogenize the sample using a homogenizer at 45 Hz for 60 seconds. Place the homogenized sample on ice for 5 minutes to completely separate nucleoprotein complexes.
1.3 Add 384 μL of chloroform and 16 μL of isoamyl alcohol, cap the tube, and vigorously shake by hand for 15 seconds. Place on ice for 10 minutes, then centrifuge at 12,000 × g for 8 minutes at 4°C. After centrifugation, the sample will separate into three layers: a red phenol-chloroform phase at the bottom, a middle layer, and a colorless aqueous phase on top. RNA is present in the aqueous phase.
1.4 Transfer approximately 500 μL of the upper aqueous phase to another RNase-free 1.5 mL EP tube, add 500 μL of isopropanol, and place at -20°C for 10 minutes, then centrifuge at 12,000 × g for 10 minutes at 4°C. (Before centrifugation, a flocculent gel-like precipitate can be observed on the side wall and bottom of the tube, which is the RNA precipitate.)
1.5 Discard the supernatant, add 1 mL of 70% RNase-free ethanol (prepared by mixing 70 mL of anhydrous ethanol with 30 mL of RNase-free water) to wash the RNA precipitate, vortex to mix, and centrifuge at 12,000 × g for 5 minutes at 4°C.
1.6 Discard the supernatant, place the tube in a laminar flow hood for 5-10 minutes to dry the RNA precipitate. (Note: Do not completely dry the RNA precipitate, as this will greatly reduce its solubility.) Resuspend the RNA precipitate in 30 μL of RNase-free water, vortex thoroughly, and let it stand for 10 minutes. Measure the concentration (OD260/280 ratio should be between 1.8-2.0) and store at -80°C.
References:Chomczynski, P., & Sacchi, N. (2006). The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nature Protocols, 1(2), 581-585.
2.Plant RNA Extraction Using a Kit Method (e.g., RNAsimple Total RNA Extraction Kit, TIANGEN:DP419)
Reagents:
a.Buffer RZ
b.Chloroform
c.Anhydrous Ethanol
d.Protein Removal Liquid RD
e.Wash Liquid RW
f.RNase-Free ddH2O
Equipment:
a.2 mL Centrifuge Tubes
b.RNase-Free Columns CR3 set
c.Homogenizer
d.Centrifuge
e.Laminar Flow Hood
f.UV-Vis Spectrophotometer for measuring OD260/280
Plant RNA Extraction
2.1 Sterilize a steel bead by high temperature and pressure, then add it to a 2 mL centrifuge tube free of RNase. Add approximately 0.2 g of plant tissue to be studied. Subsequently, add 1 mL of Buffer RZ and homogenize the sample using a homogenizer.
2.2 Allow the homogenized sample to stand at room temperature for 5 minutes to ensure complete separation of nucleoprotein complexes.
2.3 Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 5 minutes. Transfer the supernatant to a new 1.5mL RNase-free EP tube.
2.4 Add 200 μL of chloroform, cap the tube, and vortex vigorously for 15 seconds. Let the sample stand at room temperature for 3 minutes.
2.5 Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 10 minutes. The sample will separate into three layers: a yellow organic phase, an intermediate layer, and a colorless aqueous phase. RNA is primarily in the aqueous phase, which constitutes approximately 50% of the volume of the lysis buffer RZ used. Transfer the aqueous phase to a new 1.5mL RNase-free EP tube for further processing.
2.6 Slowly add 0.5 volumes of anhydrous ethanol and mix well (a precipitate may form at this stage). Transfer the solution and precipitate to the RNase-Free Columns CR3 set. Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 30 seconds. If the entire solution and mixture cannot be transferred to the RNase-Free Columns CR3 set at once, perform the transfer in two steps, centrifuging for 30 seconds at 4°C, 12,000 rpm (~13,400×g) each time, discarding the waste in the collection tube.
2.6 Add 500 μL of protein removal liquid RD (ensure ethanol has been added prior to use) to the RNase-Free Columns CR3 set. Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 30 seconds and discard the waste. Place the RNase-Free Columns CR3 set into a collection tube.
2.7 Add 500 μL of wash liquid RW (ensure ethanol has been added prior to use) to the adsorption column CR3. Let stand at room temperature for 2 minutes. Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 30 seconds and discard the waste.
2.8 Repeat step 2.7.
2.9 Place the adsorption column into a 2 mL collection tube and centrifuge at 4°C, 12,000 rpm (~13,400×g) for 2 minutes to remove residual liquid.
Note: The purpose of this step is to remove residual wash liquid from the adsorption column. After centrifugation, place the adsorption column at room temperature for a moment or in a laminar flow hood to dry thoroughly.
2.10 Transfer the adsorption column CR3 to a new 1.5 mL centrifuge tube, add 30-100 μL of RNase-Free water, let stand at room temperature for 2 minutes, and centrifuge at 4°C, 12,000 rpm (~13,400×g) for 2 minutes.
2.11 Repeat step 2.10. Measure the concentration (OD260/280 ratio should be between 1.8-2.0) and store at -80°C.
References:
Chomczynski, P., & Sacchi, N. (2006). The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nature Protocols, 1(2), 581-585.
RNAsimple Total RNA Extraction Kit DP419 User Manual.
3.Synthesis of cDNA (e.g., using PrimeScript™ RT Reagent Kit with gDNA Eraser, Takara: RR047A)
Reagents:
a.PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara: RR047A)
b.5× gDNA Eraser Buffer
c.gDNA Eraser
d.RNase Free water
e.PrimeScript RT Enzyme Mix I
f.RT Primer Mix
g.5× PrimeScript Buffer 2 (for Real Time)
Equipment:
a.Temperature-controlled PCR machine
3.1 Genomic DNA Removal Reaction:Prepare the reaction mixture on ice with the following components. To ensure the accuracy of the reaction mixture preparation, prepare the Master Mix in a quantity that is 2 more than the number of reactions, then aliquot into individual reaction tubes, and finally add the RNA sample.
Reagents Volume
5×gDNA Eraser Buffer 2.0 μl
gDNA Eraser 1.0 μl
Total RNA <1μg
RNase Free dH2O up to 10 μl
Subsequently, use a temperature-controlled PCR machine with a program set at 42°C for 2 minutes, and then quickly transfer to ice.
3.2 Reverse Transcription Reaction:Prepare the reaction mixture on ice. To ensure the accuracy of the reaction mixture preparation, prepare the Master Mix in a quantity that is 2 more than the number of reactions, then aliquot 10 μl into each reaction tube. Gently mix and immediately proceed with the reverse transcription reaction.
Reagents Volume
Reaction mixture from Step 3.1 10.0 μl
PrimeScript RT Enzyme Mix I 1.0 μl
RT Primer Mix 4.0 μl
5×PrimeScript Buffer 2(for Real Time) 4.0 μl
RNase Free dH2O 1.0 μl
Total 20 μl
3.3 Subsequently, use a temperature-controlled PCR machine with the following program:
37°C for 30 minutes
85°C for 5 seconds
Afterward, quickly transfer to ice.
References:
For a detailed understanding of the methodology and application of the PrimeScript™ RT Reagent Kit with gDNA Eraser, please refer to the following:
1. Takara Bio Inc. (2019). PrimeScript™ RT Reagent Kit with gDNA Eraser. Technical Manual. [Accessed on:2024.10.1]
4. Cloning of Fungal Effector Protein Gene A and Plant Receptor Protein Gene B
List of Reagents and Equipment Used:
a.KOD FX DNA Polymerase (TOYOBO KFX-101)
b.2x PCR buffer
c.2mM dNTPs
d.A-F and A-R Primers
e.B-F and B-R Primers
f.Fungal cDNA
g.Plant cDNA
h.ddH2O (double-distilled water)
i.Temperature-controlled PCR machine
4.1 Identification and Synthesis of Primers First, determine the nucleic acid sequences of the fungal effector protein gene A and the plant receptor protein gene B through literature review or previous research. Based on these sequences, synthesize forward and reverse primers with adapters as provided in this protocol (final concentration of 10 pmol when used):
A-F: ggcgcgccactagtg + the first 20 bp of the fungal effector protein gene A
A-R: tcccgggagcggtac + the last 20 bp of the fungal effector protein gene A (needs to be reverse complementary)
B-F: ggcgcgccactagtg + the first 20 bp of the plant receptor protein gene B
B-R: tcccgggagcggtac + the last 20 bp of the plant receptor protein gene B (needs to be reverse complementary)
4.2 PCR Amplification System for Fungal Effector Protein Gene A and Plant Receptor Protein Gene B with Adapters (using KOD FX, TOYOBO KFX-101) Before preparing the reaction mixture, thoroughly mix all reagents except for the KOD FX (enzyme solution). Completely thaw frozen reagents on ice before use.
Reagents Volume
2x PCR buffer 25 μl
2mM dNTPs 10 μl
A-F Primer 1.5 μl
A-R Primer 1.5 μl
Fungal cDNA 0.2 μg
KOD FX (1.0U/μl) 1 μl
ddH2O up to 50 μl
Reagents Volume
2x PCR buffer 25 μl
2mM dNTPs 10 μl
B-F Primer 1.5 μl
B-R Primer 1.5 μl
Plant cDNA 0.2 μg
KOD FX (1.0U/μl) 1 μl
ddH2O up to 50 μl
Add KOD FX (enzyme solution) last, vortex the reaction mixture thoroughly, spin down before proceeding with PCR.
4.3 PCR Amplification of Fungal Effector Protein Gene A and Plant Receptor Protein Gene B with Adapters Use a temperature-controlled PCR machine with the following program:
Predenature: 94°C, 2 min.
Denature: 98°C, 10 sec
Annealing: (Tm-5)°C, 30 sec
Extension: 68°C, 1kb/min
Set Denature to Extension for 33 cycles
Final extension: 68°C, 7 min.
After the reaction is complete, store the product at 4°C for future use.
References:
For detailed information on the KOD FX DNA Polymerase and its application in PCR, refer to the following:
Toyobo Co., Ltd. (2019). KOD FX DNA Polymerase. Technical Manual. [Accessed on: 2024.10.1]
Green MR, Sambrook J. (2018). PCR Protocol for Taq DNA Polymerase. Cold Spring Harbor Protocols
5. Double-Molecule Fluorescence Complementation Vectors pSPYNE and pSPYCE Double Enzyme Digestion
List of Reagents and Equipment Used:
a.pSPYNE Plasmid DNA
b.pSPYCE Plasmid DNA
c.10 x rCutSmart buffer
d.BamHI-HF restriction enzyme
e.KpnI-HF restriction enzyme
f.ddH2O (double-distilled water)
g.Temperature-controlled PCR machine
5.1 Double Enzyme Digestion Reaction Setup:Retrieve the double-molecule fluorescence complementation vectors pSPYNE and pSPYCE plasmids from the -20°C freezer, and place the 10 x rCutSmart buffer on ice. Once completely thawed, prepare the following systems:
Reagents Volume
10 x rCutSmart buffer 5 μl
BamHI-HF 1 μl
Kpn1-HF 1 μl
pSYNE Plasmid DNA 1 μg
ddH2O up to 50 μl
10 x rCutSmart buffer 5 μl
BamHI-HF 1 μl
Reagents Volume
10 x rCutSmart buffer 5 μl
BamHI-HF 1 μl
Kpn1-HF 1 μl
pSYCE Plasmid DNA 1 μg
ddH2O up to 50 μl
10 x rCutSmart buffer 5 μl
BamHI-HF 1 μl
5.2 Subsequently, use a temperature-controlled PCR machine with the following program:
37°C for 45 minutes
65°C for 45 seconds
After the reaction is complete, transfer the products to a 4°C refrigerator for storage.
References:
For detailed information on the use of restriction enzymes and plasmid DNA digestion, refer to the following:
New England Biolabs (2023). rCutSmart Buffer and HF Enzymes Technical Manual. [Accessed on: 2024.10.3]
Sambrook J, Russell DW. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
Supplementary: pSPYNE and pSPYCE vector information
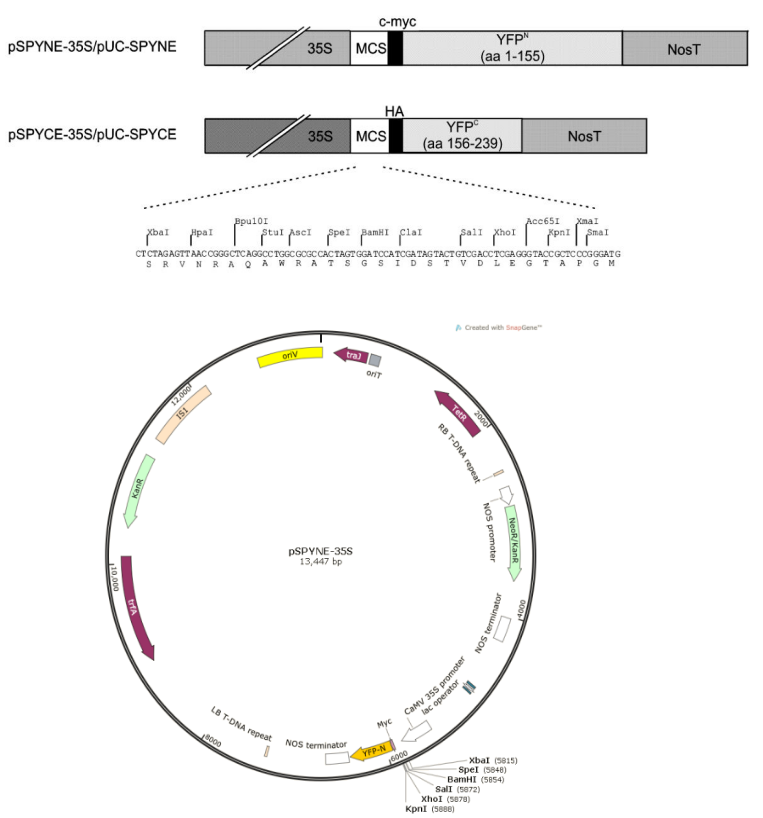
>pSPYCE vector sequence
TGAGCGTCGCAAAGGCGCTCGGTCTTGCCTTGCTCGTCGGTGATGTACTTCACCAGCTCCGCGAAGTCGCTCTTCTTGATGGAGCGCATGGGGACGTGCTTGGCAATCACGCGCACCCCCCGGCCGTTTTAGCGGCTAAAAAAGTCATGGCTCTGCCCTCGGGCGGACCACGCCCATCATGACCTTGCCAAGCTCGTCCTGCTTCTCTTCGATCTTCGCCAGCAGGGCGAGGATCGTGGCATCACCGAACCGCGCCGTGCGCGGGTCGTCGGTGAGCCAGAGTTTCAGCAGGCCGCCCAGGCGGCCCAGGTCGCCATTGATGCGGGCCAGCTCGCGGACGTGCTCATAGTCCACGACGCCCGTGATTTTGTAGCCCTGGCCGACGGCCAGCAGGTAGGCCGACAGGCTCATGCCGGCCGCCGCCGCCTTTTCCTCAATCGCTCTTCGTTCGTCTGGAAGGCAGTACACCTTGATAGGTGGGCTGCCCTTCCTGGTTGGCTTGGTTTCATCAGCCATCCGCTTGCCCTCATCTGTTACGCCGGCGGTAGCCGGCCAGCCTCGCAGAGCAGGATTCCCGTTGAGCACCGCCAGGTGCGAATAAGGGACAGTGAAGAAGGAACACCCGCTCGCGGGTGGGCCTACTTCACCTATCCTGCCCGGCTGACGCCGTTGGATACACCAAGGAAAGTCTACACGAACCCTTTGGCAAAATCCTGTATATCGTGCGAAAAAGGATGGATATACCGAAAAAATCGCTATAATGACCCCGAAGCAGGGTTATGCAGCGGAAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGTATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAGAGCGCCAGAAGGCCGCCAGAGAGGCCGAGCGCGGCCGTGAGGCTTGGACGCTAGGGCAGGGCATGAAAAAGCCCGTAGCGGGCTGCTACGGGCGTCTGACGCGGTGGAAAGGGGGAGGGGATGTTGTCTACATGGCTCTGCTGTAGTGAGTGGGTTGCGCTCCGGCAGCGGTCCTGATCAATCGTCACCCTTTCTCGGTCCTTCAACGTTCCTGACAACGAGCCTCCTTTTCGCCAATCCATCGACAATCACCGCGAGTCCCTGCTCGAACGCTGCGTCCGGACCGGCTTCGTCGAAGGCGTCTATCGCGGCCCGCAACAGCGGCGAGAGCGGAGCCTGTTCAACGGTGCCGCCGCGCTCGCCGGCATCGCTGTCGCCGGCCTGCTCCTCAAGCACGGCCCCAACAGTGAAGTAGCTGATTGTCATCAGCGCATTGACGGCGTCCCCGGCCGAAAAACCCGCCTCGCAGAGGAAGCGAAGCTGCGCGTCGGCCGTTTCCATCTGCGGTGCGCCCGGTCGCGTGCCGGCATGGATGCGCGCGCCATCGCGGTAGGCGAGCAGCGCCTGCCTGAAGCTGCGGGCATTCCCGATCAGAAATGAGCGCCAGTCGTCGTCGGCTCTCGGCACCGAATGCGTATGATTCTCCGCCAGCATGGCTTCGGCCAGTGCGTCGAGCAGCGCCCGCTTGTTCCTGAAGTGCCAGTAAAGCGCCGGCTGCTGAACCCCCAACCGTTCCGCCAGTTTGCGTGTCGTCAGACCGTCTACGCCGACCTCGTTCAACAGGTCCAGGGCGGCACGGATCACTGTATTCGGCTGCAACTTTGTCATGCTTGACACTTTATCACTGATAAACATAATATGTCCACCAACTTATCAGTGATAAAGAATCCGCGCGTTCAATCGGACCAGCGGAGGCTGGTCCGGAGGCCAGACGTGAAACCCAACATACCCCTGATCGTAATTCTGAGCACTGTCGCGCTCGACGCTGTCGGCATCGGCCTGATTATGCCGGTGCTGCCGGGCCTCCTGCGCGATCTGGTTCACTCGAACGACGTCACCGCCCACTATGGCATTCTGCTGGCGCTGTATGCGTTGGTGCAATTTGCCTGCGCACCTGTGCTGGGCGCGCTGTCGGATCGTTTCGGGCGGCGGCCAATCTTGCTCGTCTCGCTGGCCGGCGCCAGATCTGGGGAACCCTGTGGTTGGCATGCACATACAAATGGACGAACGGATAAACCTTTTCACGCCCTTTTAAATATCCGATTATTCTAATAAACGCTCTTTTCTCTTAGGTTTACCCGCCAATATATCCTGTCAAACACTGATAGTTTAAACTGAAGGCGGGAAACGACAATCTGATCATGAGCGGAGAATTAAGGGAGTCACGTTATGACCCCCGCCGATGACGCGGGACAAGCCGTTTTACGTTTGGAACTGACAGAACCGCAACGTTGAAGGAGCCACTCAGCCGCGGGTTTCTGGAGTTTAATGAGCTAAGCACATACGTCAGAAACCATTATTGCGCGTTCAAAAGTCGCCTAAGGTCACTATCAGCTAGCAAATATTTCTTGTCAAAAATGCTCCACTGACGTTCCATAAATTCCCCTCGGTATCCAATTAGAGTCTCATATTCACTCTCAATCCAAATAATCTGCACCGGATCTGGATCGTTTCGCATGATTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATGAACTGCAGGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGGAAGCCGGTCTTGTCGATCAGGATGATCTGGACGAAGAGCATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAGGCGCGCATGCCCGACGGCGATGATCTCGTCGTGACCCATGGCGATGCCTGCTTGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAGCTTGGCGGCGAATGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGAGCGGGACTCTGGGGTTCGAAATGACCGACCAAGCGACGCCCAACCTGCCATCACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTCGGAATCGTTTTCCGGGACGCCGGCTGGATGATCCTCCAGCGCGGGGATCTCATGCTGGAGTTCTTCGCCCACGGGATCTCTGCGGAACAGGCGGTCGAAGGTGCCGATATCATTACGACAGCAACGGCCGACAAGCACAACGCCACGATCCTGAGCGACAATATGATCGGGCCCGGCGTCCACATCAACGGCGTCGGCGGCGACTGCCCAGGCAAGACCGAGATGCACCGCGATATCTTGCTGCGTTCGGATATTTTCGTGGAGTTCCCGCCACAGACCCGGATGATCCCCGATCGTTCAAACATTTGGCAATAAAGTTTCTTAAGATTGAATCCTGTTGCCGGTCTTGCGATGATTATCATATAATTTCTGTTGAATTACGTTAAGCATGTAATAATTAACATGTAATGCATGACGTTATTTATGAGATGGGTTTTTATGATTAGAGTCCCGCAATTATACATTTAATACGCGATAGAAAACAAAATATAGCGCGCAAACTAGGATAAATTATCGCGCGCGGTGTCATCTATGTTACTAGATCGGGCCTCCTGTCAATGCTGGCGGCGGCTCTGGTGGTGGTTCTGGTGGCGGCTCTGAGGGTGGTGGCTCTGAGGGTGGCGGTTCTGAGGGTGGCGGCTCTGAGGGAGGCGGTTCCGGTGGTGGCTCTGGTTCCGGTGATTTTGATTATGAAAAGATGGCAAACGCTAATAAGGGGGCTATGACCGAAAATGCCGATGAAAACGCGCTACAGTCTGACGCTAAAGGCAAACTTGATTCTGTCGCTACTGATTACGGTGCTGCTATCGATGGTTTCATTGGTGACGTTTCCGGCCTTGCTAATGGTAATGGTGCTACTGGTGATTTTGCTGGCTCTAATTCCCAAATGGCTCAAGTCGGTGACGGTGATAATTCACCTTTAATGAATAATTTCCGTCAATATTTACCTTCCCTCCCTCAATCGGTTGAATGTCGCCCTTTTGTCTTTGGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGAGCGCAACGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTTGCATGCCTGCAGGTCCCCAGATTAGCCTTTTCAATTTCAGAAAGAATGCTAACCCACAGATGGTTAGAGAGGCTTACGCAGCAGGTCTCATCAAGACGATCTACCCGAGCAATAATCTCCAGGAAATCAAATACCTTCCCAAGAAGGTTAAAGATGCAGTCAAAAGATTCAGGACTAACTGCATCAAGAACACAGAGAAAGATATATTTCTCAAGATCAGAAGTACTATTCCAGTATGGACGATTCAAGGCTTGCTTCACAAACCAAGGCAAGTAATAGAGATTGGAGTCTCTAAAAAGGTAGTTCCCACTGAATCAAAGGCCATGGAGTCAAAGATTCAAATAGAGGACCTAACAGAACTCGCCGTAAAGACTGGCGAACAGTTCATACAGAGTCTCTTACGACTCAATGACAAGAAGAAAATCTTCGTCAACATGGTGGAGCACGACACACTTGTCTACTCCAAAAATATCAAAGATACAGTCTCAGAAGACCAAAGGGCAATTGAGACTTTTCAACAAAGGGTAATATCCGGAAACCTCCTCGGATTCCATTGCCCAGCTATCTGTCACTTTATTGTGAAGATAGTGGAAAAGGAAGGTGGCTCCTACAAATGCCATCATTGCGATAAAGGAAAGGCCATCGTTGAAGATGCCTCTGCCGACAGTGGTCCCAAAGATGGACCCCCACCCACGAGGAGCATCGTGGAAAAAGAAGACGTTCCAACCACGTCTTCAAAGCAAGTGGATTGATGTGATATCTCCACTGACGTAAGGGATGACGCACAATCCCACTATCCTTCGCAAGACCCTTCCTCTATATAAGGAAGTTCATTTCATTTGGAGAGAACACGGGGGACTCTAGAGTTAACCGGGCTCAGGCCTGGCGCGCCACTAGTGGATCCATCGATAGTACTGTCGACCTCGAGGGTACCGCTCCCGGGATGTACCCATACGATGTTCCAGATTACGCTGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCTACCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGTAAGAGCTCGAATTTCCCCGATCGTTCAAACATTTGGCAATAAAGTTTCTTAAGATTGAATCCTGTTGCCGGTCTTGCGATGATTATCATATAATTTCTGTTGAATTACGTTAAGCATGTAATAATTAACATGTAATGCATGACGTTATTTATGAGATGGGTTTTTATGATTAGAGTCCCGCAATTATACATTTAATACGCGATAGAAAACAAAATATAGCGCGCAAACTAGGATAAATTATCGCGCGCGGTGTCATCTATGTTACTAGATCGGGAATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGCCCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTTAGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTTGGGTGATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGGCTATTCTTTTGATTTATAAGGGATTTTGCCGATTTCGGAACCACCATCAAACAGGATTTTCGCCTGCTGGGGCAAACCAGCGTGGACCGCTTGCTGCAACTCTCTCAGGGCCAGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGGTGAAAAGAAAAACCACCCCAGTACATTAAAAACGTCCGCAATGTGTTATTAAGTTGTCTAAGCGTCAATTTGTTTACACCACAATATATCCTGCCACCAGCCAGCCAACAGCTCCCCGACCGGCAGCTCGGCACAAAATCACCACTCGATACAGGCAGCCCATCAGTCCGGGACGGCGTCAGCGGGAGAGCCGTTGTAAGGCGGCAGACTTTGCTCATGTTACCGATGCTATTCGGAAGAACGGCAACTAAGCTGCCGGGTTTGAAACACGGATGATCTCGCGGAGGGTAGCATGTTGATTGTAACGATGACAGAGCGTTGCTGCCTGTGATCAAATATCATCTCCCTCGCAGAGATCCGAATTATCAGCCTTCTTATTCATTTCTCGCTTAACCGTGACAGGCTGTCGATCTTGAGAACTATGCCGACATAATAGGAAATCGCTGGATAAAGCCGCTGAGGAAGCTGAGTGGCGCTATTTCTTTAGAAGTGAACGTTGACGATATCAACTCCCCTATCCATTGCTCACCGAATGGTACAGGTCGGGGACCCGAAGTTCCGACTGTCGGCCTGATGCATCCCCGGCTGATCGACCCCAGATCTGGGGCTGAGAAAGCCCAGTAAGGAAACAACTGTAGGTTCGAGTCGCGAGATCCCCCGGAACCAAAGGAAGTAGGTTAAACCCGCTCCGATCAGGCCGAGCCACGCCAGGCCGAGAACATTGGTTCCTGTAGGCATCGGGATTGGCGGATCAAACACTAAAGCTACTGGAACGAGCAGAAGTCCTCCGGCCGCCAGTTGCCAGGCGGTAAAGGTGAGCAGAGGCACGGGAGGTTGCCACTTGCGGGTCAGCACGGTTCCGAACGCCATGGAAACCGCCCCCGCCAGGCCCGCTGCGACGCCGACAGGATCTAGCGCTGCGTTTGGTGTCAACACCAACAGCGCCACGCCCGCAGTTCCGCAAATAGCCCCCAGGACCGCCATCAATCGTATCGGGCTACCTAGCAGAGCGGCAGAGATGAACACGACCATCAGCGGCTGCACAGCGCCTACCGTCGCCGCGACCCCGCCCGGCAGGCGGTAGACCGAAATAAACAACAAGCTCCAGAATAGCGAAATATTAAGTGCGCCGAGGATGAAGATGCGCATCCACCAGATTCCCGTTGGAATCTGTCGGACGATCATCACGAGCAATAAACCCGCCGGCAACGCCCGCAGCAGCATACCGGCGACCCCTCGGCCTCGCTGTTCGGGCTCCACGAAAACGCCGGACAGATGCGCCTTGTGAGCGTCCTTGGGGCCGTCCTCCTGTTTGAAGACCGACAGCCCAATGATCTCGCCGTCGATGTAGGCGCCGAATGCCACGGCATCTCGCAACCGTTCAGCGAACGCCTCCATGGGCTTTTTCTCCTCGTGCTCGTAAACGGACCCGAACATCTCTGGAGCTTTCTTCAGGGCCGACAATCGGATCTCGCGGAAATCCTGCACGTCGGCCGCTCCAAGCCGTCGAATCTGAGCCTTAATCACAATTGTCAATTTTAATCCTCTGTTTATCGGCAGTTCGTAGAGCGCGCCGTGCGTCCCGAGCGATACTGAGCGAAGCAAGTGCGTCGAGCAGTGCCCGCTTGTTCCTGAAATGCCAGTAAAGCGCTGGCTGCTGAACCCCCAGCCGGAACTGACCCCACAAGGCCCTAGCGTTTGCAATGCACCAGGTCATCATTGACCCAGGCGTGTTCCACCAGGCCGCTGCCTCGCAACTCTTCGCAGGCTTCGCCGACCTGCTCGCGCCACTTCTTCACGCGGGTGGAATCCGATCCGCACATGAGGCGGAAGGTTTCCAGCTTGAGCGGGTACGGCTCCCGGTGCGAGCTGAAATAGTCGAACATCCGTCGGGCCGTCGGCGACAGCTTGCGGTACTTCTCCCATATGAATTTCGTGTAGTGGTCGCCAGCAAACAGCACGACGATTTCCTCGTCGATCAGGACCTGGCAACGGGACGTTTTCTTGCCACGGTCCAGGACGCGGAAGCGGTGCAGCAGCGACACCGATTCCAGGTGCCCAACGCGGTCGGACGTGAAGCCCATCGCCGTCGCCTGTAGGCGCGACAGGCATTCCTCGGCCTTCGTGTAATACCGGCCATTGATCGACCAGCCCAGGTCCTGGCAAAGCTCGTAGAACGTGAAGGTGATCGGCTCGCCGATAGGGGTGCGCTTCGCGTACTCCAACACCTGCTGCCACACCAGTTCGTCATCGTCGGCCCGCAGCTCGACGCCGGTGTAGGTGATCTTCACGTCCTTGTTGACGTGGAAAATGACCTTGTTTTGCAGCGCCTCGCGCGGGATTTTCTTGTTGCGCGTGGTGAACAGGGCAGAGCGGGCCGTGTCGTTTGGCATCGCTCGCATCGTGTCCGGCCACGGCGCAATATCGAACAAGGAAAGCTGCATTTCCTTGATCTGCTGCTTCGTGTGTTTCAGCAACGCGGCCTGCTTGGCCTCGCTGACCTGTTTTGCCAGGTCCTCGCCGGCGGTTTTTCGCTTCTTGGTCGTCATAGTTCCTCGCGTGTCGATGGTCATCGACTTCGCCAAACCTGCCGCCTCCTGTTCGAGACGACGCGAACGCTCCACGGCGGCCGATGGCGCGGGCAGGGCAGGGGGAGCCAGTTGCACGCTGTCGCGCTCGATCTTGGCCGTAGCTTGCTGGACCATCGAGCCGACGGACTGGAAGGTTTCGCGGGGCGCACGCATGACGGTGCGGCTTGCGATGGTTTCGGCATCCTCGGCGGAAAACCCCGCGTCGATCAGTTCTTGCCTGTATGCCTTCCGGTCAAACGTCCGATTCATTCACCCTCCTTGCGGGATTGCCCCGACTCACGCCGGGGCAATGTGCCCTTATTCCTGATTTGACCCGCCTGGTGCCTTGGTGTCCAGATAATCCACCTTATCGGCAATGAAGTCGGTCCCGTAGACCGTCTGGCCGTCCTTCTCGTACTTGGTATTCCGAATCTTGCCCTGCACGAATACCAGCGACCCCTTGCCCAAATACTTGCCGTGGGCCTCGGCCTGAGAGCCAAAACACTTGATGCGGAAGAAGTCGGTGCGCTCCTGCTTGTCGCCGGCATCGTTGCGCCACATCTAGGTACTAAAACAATTCATCCAGTAAAATATAATATTTTATTTTCTCCCAATCAGGCTTGATCCCCAGTAAGTCAAAAAATAGCTCGACATACTGTTCTTCCCCGATATCCTCCCTGATCGACCGGACGCAGAAGGCAATGTCATACCACTTGTCCGCCCTGCCGCTTCTCCCAAGATCAATAAAGCCACTTACTTTGCCATCTTTCACAAAGATGTTGCTGTCTCCCAGGTCGCCGTGGGAAAAGACAAGTTCCTCTTCGGGCTTTTCCGTCTTTAAAAAATCATACAGCTCGCGCGGATCTTTAAATGGAGTGTCTTCTTCCCAGTTTTCGCAATCCACATCGGCCAGATCGTTATTCAGTAAGTAATCCAATTCGGCTAAGCGGCTGTCTAAGCTATTCGTATAGGGACAATCCGATATGTCGATGGAGTGAAAGAGCCTGATGCACTCCGCATACAGCTCGATAATCTTTTCAGGGCTTTGTTCATCTTCATACTCTTCCGAGCAAAGGACGCCATCGGCCTCACTCATGAGCAGATTGCTCCAGCCATCATGCCGTTCAAAGTGCAGGACCTTTGGAACAGGCAGCTTTCCTTCCAGCCATAGCATCATGTCCTTTTCCCGTTCCACATCATAGGTGGTCCCTTTATACCGGCTGTCCGTCATTTTTAAATATAGGTTTTCATTTTCTCCCACCAGCTTATATACCTTAGCAGGAGACATTCCTTCCGTATCTTTTACGCAGCGGTATTTTTCGATCAGTTTTTTCAATTCCGGTGATATTCTCATTTTAGCCATTTATTATTTCCTTCCTCTTTTCTACAGTATTTAAAGATACCCCAAGAAGCTAATTATAACAAGACGAACTCCAATTCACTGTTCCTTGCATTCTAAAACCTTAAATACCAGAAAACAGCTTTTTCAAAGTTGTTTTCAAAGTTGGCGTATAACATAGTATCGACGGAGCCGATTTTGAAACCACAATTATGGGTGATGCTGCCAACTTACTGATTTAGTGTATGATGGTGTTTTTGAGGTGCTCCAGTGGCTTCTGTGTCTATCAGCTGTCCCTCCTGTTCAGCTACTGACGGGGTGGTGCGTAACGGCAAAAGCACCGCCGGACATCAGCGCTATCTCTGCTCTCACTGCCGTAAAACATGGCAACTGCAGTTCACTTACACCGCTTCTCAACCCGGTACGCACCAGAAAATCATTGATATGGCCATGAATGGCGTTGGATGCCGGGCAACAGCCCGCATTATGGGCGTTGGCCTCAACACGATTTTACGTCACTTAAAAAACTCAGGCCGCAGTCGGTAACCTCGCGCATACAGCCGGGCAGTGACGTCATCGTCTGCGCGGAAATGGACGAACAGTGGGGCTATGTCGGGGCTAAATCGCGCCAGCGCTGGCTGTTTTACGCGTATGACAGTCTCCGGAAGACGGTTGTTGCGCACGTATTCGGTGAACGCACTATGGCGACGCTGGGGCGTCTTATGAGCCTGCTGTCACCCTTTGACGTGGTGATATGGATGACGGATGGCTGGCCGCTGTATGAATCCCGCCTGAAGGGAAAGCTGCACGTAATCAGCAAGCGATATACGCAGCGAATTGAGCGGCATAACCTGAATCTGAGGCAGCACCTGGCACGGCTGGGACGGAAGTCGCTGTCGTTCTCAAAATCGGTGGAGCTGCATGACAAAGTCATCGGGCATTATCTGAACATAAAACACTATCAATAAGTTGGAGTCATTACCCAATTATGATAGAATTTACAAGCTATAAGGTTATTGTCCTGGGTTTCAAGCATTAGTCCATGCAAGTTTTTATGCTTTGCCCATTCTATAGATATATTGATAAGCGCGCTGCCTATGCCTTGCCCCCTGAAATCCTTACATACGGCGATATCTTCTATATAAAAGATATATTATCTTATCAGTATTGTCAATATATTCAAGGCAATCTGCCTCCTCATCCTCTTCATCCTCTTCGTCTTGGTAGCTTTTTAAATATGGCGCTTCATAGAGTAATTCTGTAAAGGTCCAATTCTCGTTTTCATACCTCGGTATAATCTTACCTATCACCTCAAATGGTTCGCTGGGTTTATCGCACCCCCGAACACGAGCACGGCACCCGCGACCACTATGCCAAGAATGCCCAAGGTAAAAATTGCCGGCCCCGCCATGAAGTCCGTGAATGCCCCGACGGCCGAAGTGAAGGGCAGGCCGCCACCCAGGCCGCCGCCCTCACTGCCCGGCACCTGGTCGCTGAATGTCGATGCCAGCACCTGCGGCACGTCAATGCTTCCGGGCGTCGCGCTCGGGCTGATCGCCCATCCCGTTACTGCCCCGATCCCGGCAATGGCAAGGACTGCCAGCGCTGCCATTTTTGGGGTGAGGCCGTTCGCGGCCGAGGGGCGCAGCCCCTGGGGGGATGGGAGGCCCGCGTTAGCGGGCCGGGAGGGTTCGAGAAGGGGGGGCACCCCCCTTCGGCGTGCGCGGTCACGCGCACAGGGCGCAGCCCTGGTTAAAAACAAGGTTTATAAATATTGGTTTAAAAGCAGGTTAAAAGACAGGTTAGCGGTGGCCGAAAAACGGGCGGAAACCCTTGCAAATGCTGGATTTTCTGCCTGTGGACAGCCCCTCAAATGTCAATAGGTGCGCCCCTCATCTGTCAGCACTCTGCCCCTCAAGTGTCAAGGATCGCGCCCCTCATCTGTCAGTAGTCGCGCCCCTCAAGTGTCAATACCGCAGGGCACTTATCCCCAGGCTTGTCCACATCATCTGTGGGAAACTCGCGTAAAATCAGGCGTTTTCGCCGATTTGCGAGGCTGGCCAGCTCCACGTCGCCGGCCGAAATCGAGCCTGCCCCTCATCTGTCAACGCCGCGCCGGGTGAGTCGGCCCCTCAAGTGTCAACGTCCGCCCCTCATCTGTCAGTGAGGGCCAAGTTTTCCGCGAGGTATCCACAACGCCGGCGGCCGCGGTGTCTCGCACACGGCTTCGACGGCGTTTCTGGCGCGTTTGCAGGGCCATAGACGGCCGCCAGCCCAGCGGCGAGGGCAACCAGCCCGG
>pSPYNE vector sequence
TGAGCGTCGCAAAGGCGCTCGGTCTTGCCTTGCTCGTCGGTGATGTACTTCACCAGCTCCGCGAAGTCGCTCTTCTTGATGGAGCGCATGGGGACGTGCTTGGCAATCACGCGCACCCCCCGGCCGTTTTAGCGGCTAAAAAAGTCATGGCTCTGCCCTCGGGCGGACCACGCCCATCATGACCTTGCCAAGCTCGTCCTGCTTCTCTTCGATCTTCGCCAGCAGGGCGAGGATCGTGGCATCACCGAACCGCGCCGTGCGCGGGTCGTCGGTGAGCCAGAGTTTCAGCAGGCCGCCCAGGCGGCCCAGGTCGCCATTGATGCGGGCCAGCTCGCGGACGTGCTCATAGTCCACGACGCCCGTGATTTTGTAGCCCTGGCCGACGGCCAGCAGGTAGGCCGACAGGCTCATGCCGGCCGCCGCCGCCTTTTCCTCAATCGCTCTTCGTTCGTCTGGAAGGCAGTACACCTTGATAGGTGGGCTGCCCTTCCTGGTTGGCTTGGTTTCATCAGCCATCCGCTTGCCCTCATCTGTTACGCCGGCGGTAGCCGGCCAGCCTCGCAGAGCAGGATTCCCGTTGAGCACCGCCAGGTGCGAATAAGGGACAGTGAAGAAGGAACACCCGCTCGCGGGTGGGCCTACTTCACCTATCCTGCCCGGCTGACGCCGTTGGATACACCAAGGAAAGTCTACACGAACCCTTTGGCAAAATCCTGTATATCGTGCGAAAAAGGATGGATATACCGAAAAAATCGCTATAATGACCCCGAAGCAGGGTTATGCAGCGGAAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGTATCCGGTAAGCGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGGGAAACGCCTGGTATCTTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCTTTGAGTGAGCTGATACCGCTCGCCGCAGCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAAGAGCGCCAGAAGGCCGCCAGAGAGGCCGAGCGCGGCCGTGAGGCTTGGACGCTAGGGCAGGGCATGAAAAAGCCCGTAGCGGGCTGCTACGGGCGTCTGACGCGGTGGAAAGGGGGAGGGGATGTTGTCTACATGGCTCTGCTGTAGTGAGTGGGTTGCGCTCCGGCAGCGGTCCTGATCAATCGTCACCCTTTCTCGGTCCTTCAACGTTCCTGACAACGAGCCTCCTTTTCGCCAATCCATCGACAATCACCGCGAGTCCCTGCTCGAACGCTGCGTCCGGACCGGCTTCGTCGAAGGCGTCTATCGCGGCCCGCAACAGCGGCGAGAGCGGAGCCTGTTCAACGGTGCCGCCGCGCTCGCCGGCATCGCTGTCGCCGGCCTGCTCCTCAAGCACGGCCCCAACAGTGAAGTAGCTGATTGTCATCAGCGCATTGACGGCGTCCCCGGCCGAAAAACCCGCCTCGCAGAGGAAGCGAAGCTGCGCGTCGGCCGTTTCCATCTGCGGTGCGCCCGGTCGCGTGCCGGCATGGATGCGCGCGCCATCGCGGTAGGCGAGCAGCGCCTGCCTGAAGCTGCGGGCATTCCCGATCAGAAATGAGCGCCAGTCGTCGTCGGCTCTCGGCACCGAATGCGTATGATTCTCCGCCAGCATGGCTTCGGCCAGTGCGTCGAGCAGCGCCCGCTTGTTCCTGAAGTGCCAGTAAAGCGCCGGCTGCTGAACCCCCAACCGTTCCGCCAGTTTGCGTGTCGTCAGACCGTCTACGCCGACCTCGTTCAACAGGTCCAGGGCGGCACGGATCACTGTATTCGGCTGCAACTTTGTCATGCTTGACACTTTATCACTGATAAACATAATATGTCCACCAACTTATCAGTGATAAAGAATCCGCGCGTTCAATCGGACCAGCGGAGGCTGGTCCGGAGGCCAGACGTGAAACCCAACATACCCCTGATCGTAATTCTGAGCACTGTCGCGCTCGACGCTGTCGGCATCGGCCTGATTATGCCGGTGCTGCCGGGCCTCCTGCGCGATCTGGTTCACTCGAACGACGTCACCGCCCACTATGGCATTCTGCTGGCGCTGTATGCGTTGGTGCAATTTGCCTGCGCACCTGTGCTGGGCGCGCTGTCGGATCGTTTCGGGCGGCGGCCAATCTTGCTCGTCTCGCTGGCCGGCGCCAGATCTGGGGAACCCTGTGGTTGGCATGCACATACAAATGGACGAACGGATAAACCTTTTCACGCCCTTTTAAATATCCGATTATTCTAATAAACGCTCTTTTCTCTTAGGTTTACCCGCCAATATATCCTGTCAAACACTGATAGTTTAAACTGAAGGCGGGAAACGACAATCTGATCATGAGCGGAGAATTAAGGGAGTCACGTTATGACCCCCGCCGATGACGCGGGACAAGCCGTTTTACGTTTGGAACTGACAGAACCGCAACGTTGAAGGAGCCACTCAGCCGCGGGTTTCTGGAGTTTAATGAGCTAAGCACATACGTCAGAAACCATTATTGCGCGTTCAAAAGTCGCCTAAGGTCACTATCAGCTAGCAAATATTTCTTGTCAAAAATGCTCCACTGACGTTCCATAAATTCCCCTCGGTATCCAATTAGAGTCTCATATTCACTCTCAATCCAAATAATCTGCACCGGATCTGGATCGTTTCGCATGATTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATGAACTGCAGGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGGAAGCCGGTCTTGTCGATCAGGATGATCTGGACGAAGAGCATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAGGCGCGCATGCCCGACGGCGATGATCTCGTCGTGACCCATGGCGATGCCTGCTTGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAGCTTGGCGGCGAATGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGAGCGGGACTCTGGGGTTCGAAATGACCGACCAAGCGACGCCCAACCTGCCATCACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTCGGAATCGTTTTCCGGGACGCCGGCTGGATGATCCTCCAGCGCGGGGATCTCATGCTGGAGTTCTTCGCCCACGGGATCTCTGCGGAACAGGCGGTCGAAGGTGCCGATATCATTACGACAGCAACGGCCGACAAGCACAACGCCACGATCCTGAGCGACAATATGATCGGGCCCGGCGTCCACATCAACGGCGTCGGCGGCGACTGCCCAGGCAAGACCGAGATGCACCGCGATATCTTGCTGCGTTCGGATATTTTCGTGGAGTTCCCGCCACAGACCCGGATGATCCCCGATCGTTCAAACATTTGGCAATAAAGTTTCTTAAGATTGAATCCTGTTGCCGGTCTTGCGATGATTATCATATAATTTCTGTTGAATTACGTTAAGCATGTAATAATTAACATGTAATGCATGACGTTATTTATGAGATGGGTTTTTATGATTAGAGTCCCGCAATTATACATTTAATACGCGATAGAAAACAAAATATAGCGCGCAAACTAGGATAAATTATCGCGCGCGGTGTCATCTATGTTACTAGATCGGGCCTCCTGTCAATGCTGGCGGCGGCTCTGGTGGTGGTTCTGGTGGCGGCTCTGAGGGTGGTGGCTCTGAGGGTGGCGGTTCTGAGGGTGGCGGCTCTGAGGGAGGCGGTTCCGGTGGTGGCTCTGGTTCCGGTGATTTTGATTATGAAAAGATGGCAAACGCTAATAAGGGGGCTATGACCGAAAATGCCGATGAAAACGCGCTACAGTCTGACGCTAAAGGCAAACTTGATTCTGTCGCTACTGATTACGGTGCTGCTATCGATGGTTTCATTGGTGACGTTTCCGGCCTTGCTAATGGTAATGGTGCTACTGGTGATTTTGCTGGCTCTAATTCCCAAATGGCTCAAGTCGGTGACGGTGATAATTCACCTTTAATGAATAATTTCCGTCAATATTTACCTTCCCTCCCTCAATCGGTTGAATGTCGCCCTTTTGTCTTTGGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGAGCGCAACGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATGTTGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTTGCATGCCTGCAGGTCCCCAGATTAGCCTTTTCAATTTCAGAAAGAATGCTAACCCACAGATGGTTAGAGAGGCTTACGCAGCAGGTCTCATCAAGACGATCTACCCGAGCAATAATCTCCAGGAAATCAAATACCTTCCCAAGAAGGTTAAAGATGCAGTCAAAAGATTCAGGACTAACTGCATCAAGAACACAGAGAAAGATATATTTCTCAAGATCAGAAGTACTATTCCAGTATGGACGATTCAAGGCTTGCTTCACAAACCAAGGCAAGTAATAGAGATTGGAGTCTCTAAAAAGGTAGTTCCCACTGAATCAAAGGCCATGGAGTCAAAGATTCAAATAGAGGACCTAACAGAACTCGCCGTAAAGACTGGCGAACAGTTCATACAGAGTCTCTTACGACTCAATGACAAGAAGAAAATCTTCGTCAACATGGTGGAGCACGACACACTTGTCTACTCCAAAAATATCAAAGATACAGTCTCAGAAGACCAAAGGGCAATTGAGACTTTTCAACAAAGGGTAATATCCGGAAACCTCCTCGGATTCCATTGCCCAGCTATCTGTCACTTTATTGTGAAGATAGTGGAAAAGGAAGGTGGCTCCTACAAATGCCATCATTGCGATAAAGGAAAGGCCATCGTTGAAGATGCCTCTGCCGACAGTGGTCCCAAAGATGGACCCCCACCCACGAGGAGCATCGTGGAAAAAGAAGACGTTCCAACCACGTCTTCAAAGCAAGTGGATTGATGTGATATCTCCACTGACGTAAGGGATGACGCACAATCCCACTATCCTTCGCAAGACCCTTCCTCTATATAAGGAAGTTCATTTCATTTGGAGAGAACACGGGGGACTCTAGAGTTAACCGGGCTCAGGCCTGGCGCGCCACTAGTGGATCCATCGATAGTACTGTCGACCTCGAGGGTACCGCTCCCGGGATGGAGCAAAAGTTGATTTCTGAGGAGGATCTTATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCTTCGGCTACGGCCTGCAGTGCTTCGCCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGCCTAAGAGCTCGAATTTCCCCGATCGTTCAAACATTTGGCAATAAAGTTTCTTAAGATTGAATCCTGTTGCCGGTCTTGCGATGATTATCATATAATTTCTGTTGAATTACGTTAAGCATGTAATAATTAACATGTAATGCATGACGTTATTTATGAGATGGGTTTTTATGATTAGAGTCCCGCAATTATACATTTAATACGCGATAGAAAACAAAATATAGCGCGCAAACTAGGATAAATTATCGCGCGCGGTGTCATCTATGTTACTAGATCGGGAATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGCCCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTTAGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTTGGGTGATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGGCTATTCTTTTGATTTATAAGGGATTTTGCCGATTTCGGAACCACCATCAAACAGGATTTTCGCCTGCTGGGGCAAACCAGCGTGGACCGCTTGCTGCAACTCTCTCAGGGCCAGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGGTGAAAAGAAAAACCACCCCAGTACATTAAAAACGTCCGCAATGTGTTATTAAGTTGTCTAAGCGTCAATTTGTTTACACCACAATATATCCTGCCACCAGCCAGCCAACAGCTCCCCGACCGGCAGCTCGGCACAAAATCACCACTCGATACAGGCAGCCCATCAGTCCGGGACGGCGTCAGCGGGAGAGCCGTTGTAAGGCGGCAGACTTTGCTCATGTTACCGATGCTATTCGGAAGAACGGCAACTAAGCTGCCGGGTTTGAAACACGGATGATCTCGCGGAGGGTAGCATGTTGATTGTAACGATGACAGAGCGTTGCTGCCTGTGATCAAATATCATCTCCCTCGCAGAGATCCGAATTATCAGCCTTCTTATTCATTTCTCGCTTAACCGTGACAGGCTGTCGATCTTGAGAACTATGCCGACATAATAGGAAATCGCTGGATAAAGCCGCTGAGGAAGCTGAGTGGCGCTATTTCTTTAGAAGTGAACGTTGACGATATCAACTCCCCTATCCATTGCTCACCGAATGGTACAGGTCGGGGACCCGAAGTTCCGACTGTCGGCCTGATGCATCCCCGGCTGATCGACCCCAGATCTGGGGCTGAGAAAGCCCAGTAAGGAAACAACTGTAGGTTCGAGTCGCGAGATCCCCCGGAACCAAAGGAAGTAGGTTAAACCCGCTCCGATCAGGCCGAGCCACGCCAGGCCGAGAACATTGGTTCCTGTAGGCATCGGGATTGGCGGATCAAACACTAAAGCTACTGGAACGAGCAGAAGTCCTCCGGCCGCCAGTTGCCAGGCGGTAAAGGTGAGCAGAGGCACGGGAGGTTGCCACTTGCGGGTCAGCACGGTTCCGAACGCCATGGAAACCGCCCCCGCCAGGCCCGCTGCGACGCCGACAGGATCTAGCGCTGCGTTTGGTGTCAACACCAACAGCGCCACGCCCGCAGTTCCGCAAATAGCCCCCAGGACCGCCATCAATCGTATCGGGCTACCTAGCAGAGCGGCAGAGATGAACACGACCATCAGCGGCTGCACAGCGCCTACCGTCGCCGCGACCCCGCCCGGCAGGCGGTAGACCGAAATAAACAACAAGCTCCAGAATAGCGAAATATTAAGTGCGCCGAGGATGAAGATGCGCATCCACCAGATTCCCGTTGGAATCTGTCGGACGATCATCACGAGCAATAAACCCGCCGGCAACGCCCGCAGCAGCATACCGGCGACCCCTCGGCCTCGCTGTTCGGGCTCCACGAAAACGCCGGACAGATGCGCCTTGTGAGCGTCCTTGGGGCCGTCCTCCTGTTTGAAGACCGACAGCCCAATGATCTCGCCGTCGATGTAGGCGCCGAATGCCACGGCATCTCGCAACCGTTCAGCGAACGCCTCCATGGGCTTTTTCTCCTCGTGCTCGTAAACGGACCCGAACATCTCTGGAGCTTTCTTCAGGGCCGACAATCGGATCTCGCGGAAATCCTGCACGTCGGCCGCTCCAAGCCGTCGAATCTGAGCCTTAATCACAATTGTCAATTTTAATCCTCTGTTTATCGGCAGTTCGTAGAGCGCGCCGTGCGTCCCGAGCGATACTGAGCGAAGCAAGTGCGTCGAGCAGTGCCCGCTTGTTCCTGAAATGCCAGTAAAGCGCTGGCTGCTGAACCCCCAGCCGGAACTGACCCCACAAGGCCCTAGCGTTTGCAATGCACCAGGTCATCATTGACCCAGGCGTGTTCCACCAGGCCGCTGCCTCGCAACTCTTCGCAGGCTTCGCCGACCTGCTCGCGCCACTTCTTCACGCGGGTGGAATCCGATCCGCACATGAGGCGGAAGGTTTCCAGCTTGAGCGGGTACGGCTCCCGGTGCGAGCTGAAATAGTCGAACATCCGTCGGGCCGTCGGCGACAGCTTGCGGTACTTCTCCCATATGAATTTCGTGTAGTGGTCGCCAGCAAACAGCACGACGATTTCCTCGTCGATCAGGACCTGGCAACGGGACGTTTTCTTGCCACGGTCCAGGACGCGGAAGCGGTGCAGCAGCGACACCGATTCCAGGTGCCCAACGCGGTCGGACGTGAAGCCCATCGCCGTCGCCTGTAGGCGCGACAGGCATTCCTCGGCCTTCGTGTAATACCGGCCATTGATCGACCAGCCCAGGTCCTGGCAAAGCTCGTAGAACGTGAAGGTGATCGGCTCGCCGATAGGGGTGCGCTTCGCGTACTCCAACACCTGCTGCCACACCAGTTCGTCATCGTCGGCCCGCAGCTCGACGCCGGTGTAGGTGATCTTCACGTCCTTGTTGACGTGGAAAATGACCTTGTTTTGCAGCGCCTCGCGCGGGATTTTCTTGTTGCGCGTGGTGAACAGGGCAGAGCGGGCCGTGTCGTTTGGCATCGCTCGCATCGTGTCCGGCCACGGCGCAATATCGAACAAGGAAAGCTGCATTTCCTTGATCTGCTGCTTCGTGTGTTTCAGCAACGCGGCCTGCTTGGCCTCGCTGACCTGTTTTGCCAGGTCCTCGCCGGCGGTTTTTCGCTTCTTGGTCGTCATAGTTCCTCGCGTGTCGATGGTCATCGACTTCGCCAAACCTGCCGCCTCCTGTTCGAGACGACGCGAACGCTCCACGGCGGCCGATGGCGCGGGCAGGGCAGGGGGAGCCAGTTGCACGCTGTCGCGCTCGATCTTGGCCGTAGCTTGCTGGACCATCGAGCCGACGGACTGGAAGGTTTCGCGGGGCGCACGCATGACGGTGCGGCTTGCGATGGTTTCGGCATCCTCGGCGGAAAACCCCGCGTCGATCAGTTCTTGCCTGTATGCCTTCCGGTCAAACGTCCGATTCATTCACCCTCCTTGCGGGATTGCCCCGACTCACGCCGGGGCAATGTGCCCTTATTCCTGATTTGACCCGCCTGGTGCCTTGGTGTCCAGATAATCCACCTTATCGGCAATGAAGTCGGTCCCGTAGACCGTCTGGCCGTCCTTCTCGTACTTGGTATTCCGAATCTTGCCCTGCACGAATACCAGCGACCCCTTGCCCAAATACTTGCCGTGGGCCTCGGCCTGAGAGCCAAAACACTTGATGCGGAAGAAGTCGGTGCGCTCCTGCTTGTCGCCGGCATCGTTGCGCCACATCTAGGTACTAAAACAATTCATCCAGTAAAATATAATATTTTATTTTCTCCCAATCAGGCTTGATCCCCAGTAAGTCAAAAAATAGCTCGACATACTGTTCTTCCCCGATATCCTCCCTGATCGACCGGACGCAGAAGGCAATGTCATACCACTTGTCCGCCCTGCCGCTTCTCCCAAGATCAATAAAGCCACTTACTTTGCCATCTTTCACAAAGATGTTGCTGTCTCCCAGGTCGCCGTGGGAAAAGACAAGTTCCTCTTCGGGCTTTTCCGTCTTTAAAAAATCATACAGCTCGCGCGGATCTTTAAATGGAGTGTCTTCTTCCCAGTTTTCGCAATCCACATCGGCCAGATCGTTATTCAGTAAGTAATCCAATTCGGCTAAGCGGCTGTCTAAGCTATTCGTATAGGGACAATCCGATATGTCGATGGAGTGAAAGAGCCTGATGCACTCCGCATACAGCTCGATAATCTTTTCAGGGCTTTGTTCATCTTCATACTCTTCCGAGCAAAGGACGCCATCGGCCTCACTCATGAGCAGATTGCTCCAGCCATCATGCCGTTCAAAGTGCAGGACCTTTGGAACAGGCAGCTTTCCTTCCAGCCATAGCATCATGTCCTTTTCCCGTTCCACATCATAGGTGGTCCCTTTATACCGGCTGTCCGTCATTTTTAAATATAGGTTTTCATTTTCTCCCACCAGCTTATATACCTTAGCAGGAGACATTCCTTCCGTATCTTTTACGCAGCGGTATTTTTCGATCAGTTTTTTCAATTCCGGTGATATTCTCATTTTAGCCATTTATTATTTCCTTCCTCTTTTCTACAGTATTTAAAGATACCCCAAGAAGCTAATTATAACAAGACGAACTCCAATTCACTGTTCCTTGCATTCTAAAACCTTAAATACCAGAAAACAGCTTTTTCAAAGTTGTTTTCAAAGTTGGCGTATAACATAGTATCGACGGAGCCGATTTTGAAACCACAATTATGGGTGATGCTGCCAACTTACTGATTTAGTGTATGATGGTGTTTTTGAGGTGCTCCAGTGGCTTCTGTGTCTATCAGCTGTCCCTCCTGTTCAGCTACTGACGGGGTGGTGCGTAACGGCAAAAGCACCGCCGGACATCAGCGCTATCTCTGCTCTCACTGCCGTAAAACATGGCAACTGCAGTTCACTTACACCGCTTCTCAACCCGGTACGCACCAGAAAATCATTGATATGGCCATGAATGGCGTTGGATGCCGGGCAACAGCCCGCATTATGGGCGTTGGCCTCAACACGATTTTACGTCACTTAAAAAACTCAGGCCGCAGTCGGTAACCTCGCGCATACAGCCGGGCAGTGACGTCATCGTCTGCGCGGAAATGGACGAACAGTGGGGCTATGTCGGGGCTAAATCGCGCCAGCGCTGGCTGTTTTACGCGTATGACAGTCTCCGGAAGACGGTTGTTGCGCACGTATTCGGTGAACGCACTATGGCGACGCTGGGGCGTCTTATGAGCCTGCTGTCACCCTTTGACGTGGTGATATGGATGACGGATGGCTGGCCGCTGTATGAATCCCGCCTGAAGGGAAAGCTGCACGTAATCAGCAAGCGATATACGCAGCGAATTGAGCGGCATAACCTGAATCTGAGGCAGCACCTGGCACGGCTGGGACGGAAGTCGCTGTCGTTCTCAAAATCGGTGGAGCTGCATGACAAAGTCATCGGGCATTATCTGAACATAAAACACTATCAATAAGTTGGAGTCATTACCCAATTATGATAGAATTTACAAGCTATAAGGTTATTGTCCTGGGTTTCAAGCATTAGTCCATGCAAGTTTTTATGCTTTGCCCATTCTATAGATATATTGATAAGCGCGCTGCCTATGCCTTGCCCCCTGAAATCCTTACATACGGCGATATCTTCTATATAAAAGATATATTATCTTATCAGTATTGTCAATATATTCAAGGCAATCTGCCTCCTCATCCTCTTCATCCTCTTCGTCTTGGTAGCTTTTTAAATATGGCGCTTCATAGAGTAATTCTGTAAAGGTCCAATTCTCGTTTTCATACCTCGGTATAATCTTACCTATCACCTCAAATGGTTCGCTGGGTTTATCGCACCCCCGAACACGAGCACGGCACCCGCGACCACTATGCCAAGAATGCCCAAGGTAAAAATTGCCGGCCCCGCCATGAAGTCCGTGAATGCCCCGACGGCCGAAGTGAAGGGCAGGCCGCCACCCAGGCCGCCGCCCTCACTGCCCGGCACCTGGTCGCTGAATGTCGATGCCAGCACCTGCGGCACGTCAATGCTTCCGGGCGTCGCGCTCGGGCTGATCGCCCATCCCGTTACTGCCCCGATCCCGGCAATGGCAAGGACTGCCAGCGCTGCCATTTTTGGGGTGAGGCCGTTCGCGGCCGAGGGGCGCAGCCCCTGGGGGGATGGGAGGCCCGCGTTAGCGGGCCGGGAGGGTTCGAGAAGGGGGGGCACCCCCCTTCGGCGTGCGCGGTCACGCGCACAGGGCGCAGCCCTGGTTAAAAACAAGGTTTATAAATATTGGTTTAAAAGCAGGTTAAAAGACAGGTTAGCGGTGGCCGAAAAACGGGCGGAAACCCTTGCAAATGCTGGATTTTCTGCCTGTGGACAGCCCCTCAAATGTCAATAGGTGCGCCCCTCATCTGTCAGCACTCTGCCCCTCAAGTGTCAAGGATCGCGCCCCTCATCTGTCAGTAGTCGCGCCCCTCAAGTGTCAATACCGCAGGGCACTTATCCCCAGGCTTGTCCACATCATCTGTGGGAAACTCGCGTAAAATCAGGCGTTTTCGCCGATTTGCGAGGCTGGCCAGCTCCACGTCGCCGGCCGAAATCGAGCCTGCCCCTCATCTGTCAACGCCGCGCCGGGTGAGTCGGCCCCTCAAGTGTCAACGTCCGCCCCTCATCTGTCAGTGAGGGCCAAGTTTTCCGCGAGGTATCCACAACGCCGGCGGCCGCGGTGTCTCGCACACGGCTTCGACGGCGTTTCTGGCGCGTTTGCAGGGCCATAGACGGCCGCCAGCCCAGCGGCGAGGGCAACCAGCCCGG
6. Agarose Gel Electrophoresis and Gel Extraction (EasyPure Quick Gel Extraction Kit, TransGen Biotech: EG101)
List of Reagents and Equipment Used:
a.Agarose
b.Gel-Red nucleic acid dye (10,000x)
c.Trans2K® Plus II DNA Marker
d.EasyPure Quick Gel Extraction Kit
e.Gel Solubilization Buffer (GSB)
f.Ethylenediaminetetraacetic acid (EDTA)
g.Glacial acetic acid
h.Wash Buffer (WB)
i.Elution Buffer (EB)
j.Tris(hydroxymethyl) aminomethane(Tris)
k.Gel Spin Columns with Collection Tubes
l.Microcentrifuge tubes
m.Microwave
n.Water bath
o.UV light
p.Centrifuge
6.1 Preparation of 1.5% Agarose Gel: Prepare a 1.5% agarose gel by adding 1.5g of agarose powder to 100mL of 1×TAE buffer (where 50xTAE: ddH2O = 1:49). Heat the mixture in a microwave until the agarose is completely dissolved, taking care to avoid excessive boiling that could lead to evaporation and affect the final concentration. Once the mixture has cooled to approximately 50°C, add 10μl of Gel-Red nucleic acid dye (10,000 x) .
6.2 Gel Casting: Pour the gel solution into a casting mold, insert a comb at the appropriate position, and allow it to solidify at room temperature for 40 minutes. Remove the comb and place the gel into the electrophoresis tank, ensuring that the wells are on the negative pole side.
6.3 Electrophoresis: Fill the tank with 1×TAE buffer to a level about 1mm above the gel surface. Load 10μL of PCR product, which has been mixed with 2μL of 6x DNA loading buffer, into each well. Add 5μL of Trans2K® Plus II DNA Marker (TransGen Biotech: BM121-01) to the leftmost well and run the electrophoresis at 120V for approximately 25 to 40 minutes .
6.4 Gel Cutting and DNA Fragment Extraction:After electrophoresis, carefully excise the gel region containing the target DNA fragment under a UV light. Place the excised gel piece into a 1.5 mL centrifuge tube and add Gel Solubilization Buffer in a volume three times that of the gel weight. Incubate at 55°C in a water bath until the gel piece is completely dissolved.
6.5 Gel Dissolution and DNA Binding:Once the dissolved gel solution has cooled to room temperature (high temperatures weaken the DNA binding to the Gel Spin Columns), add it to the Gel Spin Columns with Collection Tubes and let it sit for 1 minute. Centrifuge at 10,000×g for 1 minute and discard the flow-through.
6.6 Washing and DNA Elution: Add 650μl of Wash Buffer, centrifuge at 10,000×g for 1 minute, and discard the flow-through. Centrifuge again at 10,000×g for 1-2 minutes to thoroughly remove any residual Wash Buffer.
6.7 DNA Elution:Place the Gel Spin Columns in a clean microcentrifuge tube, let it stand for 1 minute to allow any residual ethanol to evaporate, then add 30-50 μl of deionized water to the center of the column matrix. Let it stand at room temperature for 1 minute.
6.8 DNA Collection:Centrifuge at 10,000×g for 1 minute to elute the nucleic acid. The eluted nucleic acid can be stored at 4°C.
Supplementary Information:
50x TAE (Tris-Acetate-EDTA) Buffer Recipe
Reagents Amount
Tris 242.28 g
Glacial acetic acid 60.05 g
EDTA 18.612 g
ddH2O up to 1 L
References:
For detailed procedures on agarose gel electrophoresis and gel extraction, refer to:
Sambrook J, Russell DW. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
Green MR, Sambrook J. (2018). PCR Protocol for Taq DNA Polymerase. Cold Spring Harbor Protocols.
7. Construction of pSPYNE-A and pSPYCE-B Vectors by Homologous Recombination (ClonExpress® Ultra One Step Cloning Kit, Vazyme: C115)
List of Reagents and Equipment Used:
a.Linearized vector pSPYNE
b.Insert fragment A and B
c.ClonExpress® Ultra One Step Cloning Kit
d.2 × ClonExpress Mix
e.ddH2O (double-distilled water)
f.Temperature-controlled PCR machine
g.Pipettes and tips
h.Microcentrifuge tubes
7.1 Calculation of Linearized Vector and Insert Dosage:
The optimal cloning vector dosage is calculated as follows:
[0.02 × base pairs (bp) of cloning vector] ng (0.03 pmol)
The optimal insert dosage is calculated as follows:
[0.04 × bp of insert fragment] ng (0.06 pmol)
Note: Calculate the required DNA amounts for the recombination reaction using the formulas provided. To ensure accuracy in pipetting, dilute the linearized vector and insert fragments appropriately before preparing the recombination reaction system, with each component's volume not less than 1μl.
7.2 Preparation of the Reaction System on Ice
Components Volume
Linearized vector pSPYNE X μl
Insert fragment A Y μl
2 × ClonExpress Mix 5 μl
ddH2O to 10 μl
Components Volume
Linearized vector pSPYCE X μl
Insert fragment B Y μl
2 × ClonExpress Mix 5 μl
ddH2O to 10 μl
Gently mix by pipetting up and down (do not vortex), and briefly centrifuge to collect the reaction liquid at the bottom of the tube.
7.3 Subsequent Use of a Temperature-Controlled PCR Machine with the Following Program
50°C for 30 minutes
After the reaction is complete, transfer to a 4°C refrigerator for storage.
References:
For detailed procedures on one-step cloning kits and homologous recombination, refer to:
Vazyme Technical Manual. (2019). ClonExpress® Ultra One Step Cloning Kit. [Accessed on: 2024.10.3]
Sambrook J, Russell DW. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
8. Transformation of pSPYNE-A and pSPYCE-B Recombinant Products
List of Reagents and Equipment Used:
a.Chemically competent cells (e.g., DH5α)
b.pSPYNE-A and pSPYCE-B recombinant products
c.10 x rCutSmart buffer
d.BamHI-HF and KpnI-HF restriction enzymes
e.ddH2O (double-distilled water)
f.Temperature-controlled PCR machine
g.LB liquid medium and agar plates with Kanamycin resistance
h.KOD FX DNA Polymerase (TOYOBO KFX-101)
i.2x PCR buffer
j.2mM dNTPs
k.Forward primer X-F and reverse primers A-R and B-R
l.Agarose
m.Gel-Red nucleic acid dye
n.1×TAE buffer
o.Trans2K® Plus II DNA Marker
8.1 Transformation of Chemically Competent Cells:Place the chemically competent cells used for cloning on ice to thaw (e.g., DH5α Competent Cells, Vazyme, C502).
8.2 Recombinant Product Transformation,Take 10 μl of the pSPYNE-A and pSPYCE-B recombinant products and add them separately to 100 μl of competent cells. Gently tap the tube to mix (do not vortex) and let sit on ice for 30 minutes. Note: The volume of recombinant product for transformation should not exceed 1/10 of the volume of the competent cells used.
8.3 Heat Shock:Transfer to a 42°C water bath for 45 seconds, then immediately cool on ice for 2 minutes.
8.4 Recovery in Liquid Media:Add 800 μl of LB liquid medium (without antibiotics) and incubate at 37°C with shaking at 200 rpm for 1 hour.
8.5 Pre-warming of Agar Plates:Pre-warm the LB agar plates containing Kanamycin resistance in a 37°C incubator.
8.6 Centrifugation and Plating:Centrifuge at 5,000 rpm (2,500 × g) for 5 minutes, discard the 800 μl supernatant. Resuspend the pellet in the remaining medium and gently spread with a sterile spreader on plates with the correct antibiotic resistance.
8.7 Incubation:Invert and incubate in a 37°C incubator for 12-16 hours.
8.8 Picking Single Colonies:Pick single colonies into 1.5 ml tubes containing 1 ml of LB liquid medium, add 1 μl of Kanamycin antibiotic, and incubate at 37°C with shaking at 200 rpm for 6 hours. Generally, 8-12 single clones are used for material identification.
8.9 PCR Amplification System Configuration (using KOD FX, TOYOBO KFX-101)
Both pSPYNE-A and pSPYCE-B use a common forward primer X-F for PCR amplification:
X-F: 5’-AGTTAACCGGGCTCAGGCCT-3’
Mix all reagents thoroughly on ice before adding the KOD FX (enzyme solution). Ensure all frozen reagents are completely thawed on ice before use.
Reagents Volume
2x PCR buffer 10 μl
2mM dNTPs 4 μl
X-F Primer 0.6 μl
A-R Primer 0.6 μl
Bacterial culture 1ul
KOD FX (1.0U/μl) 0.4 μl
ddH2O up to 20 μl
Reagents Volume
2x PCR buffer 10 μl
2mM dNTPs 4 μl
X-F Primer 0.6 μl
B-R Primer 0.6 μl
Bacterial culture 1ul
KOD FX (1.0U/μl) 0.4 μl
ddH2O up to 20 μl
Add KOD FX (enzyme solution) last, vortex the reaction mixture thoroughly, and centrifuge before proceeding with PCR.
8.10 PCR Amplification Program:
Predenature: 94°C, 2 min.
Denature: 98°C, 10 sec
Annealing: (Tm-5)°C, 30 sec
Extension: 68°C, 1kb/min
Set Denature to Extension for 33 cycles
Final extension: 68°C, 7 min.
After the reaction is complete, transfer the products to a 4°C refrigerator for storage.
8.11 Agarose Gel Preparation:Prepare a 1.5% agarose gel by adding 1.5 g of agarose powder to 100 ml of 1×TAE buffer (i.e., 50xTAE: ddH2O = 1:49). Microwave until the agarose is completely dissolved, avoiding excessive boiling that could lead to evaporation and affect the final concentration. When cooled to approximately 50°C, add 10 μl of Gel-Red nucleic acid dye (10,000x).
8.12 Gel Casting:Pour the gel into a casting mold, insert a comb at the appropriate position, and let it solidify at room temperature for 40 minutes. Remove the comb, place the gel into the electrophoresis tank, ensuring the wells are on the negative pole side.
8.13 Electrophoresis:Fill the tank with 1×TAE to a level just covering the gel surface. Load 10 μl of PCR product mixed with 2 μl of 6x DNA loading buffer into each well. Load 5 μl of Trans2K® Plus II DNA Marker (TransGen Biotech, BM121-01) into the leftmost well. Run the electrophoresis at 120V for approximately 25-40 minutes.
8.14 Gel Visualization and Sequencing:After electrophoresis, observe the band sizes under a UV gel doc to confirm they match the expected sizes. Send the successfully amplified bacterial cultures and primers X-F, A-R, B-R for sequencing to a sequencing company.
Supplementary Information:
LB Medium (Luria-Bertani Medium) is a commonly used bacterial growth medium suitable for a variety of bacteria.
Liquid LB Medium Recipe:
Reagents Volume
Tryptone 10 g
Yeast Extract 5 g
NaCl 10 g
ddH2O up to 1 L
Note: Dissolve the above components in 1 liter of water, mix well, and adjust the pH to 7.0-7.5. Sterilize by autoclaving at 121°C for 20 minutes. The medium should appear as a uniform yellow liquid after sterilization.
Solid LB Medium Recipe:
Reagents Volume
Tryptone 10 g
Yeast Extract 5 g
Agar 7.5 g
NaCl 10 g
ddH2O up to 1 L
Note: Dissolve the above components in 1 liter of water, mix well, and adjust the pH to 7.0-7.5. Sterilize by autoclaving at 121°C for 20 minutes. After sterilization and cooling to 50-60°C, add Kanamycin antibiotic (final concentration of 50 μg/ml) under a laminar flow hood near an alcohol lamp flame and mix quickly. Pour into petri dishes and allow to solidify before use.
References:
For detailed information on competent cell preparation and transformation, refer to:
Sambrook J, Russell DW. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
For information on PCR amplification and agarose gel electrophoresis, refer to:
Green MR, Sambrook J. (2018). PCR Protocol for Taq DNA Polymerase. Cold Spring Harbor Protocols.
9. Plasmid Extraction of Recombinant Products pSPYNE-A and pSPYCE-B (TIANprep Mini Plasmid Kit, TIANGEN BIOTECH: DP103)
Note: Preserve the E. coli strains with correct sequence alignment after sequencing by mixing with 50% sterile glycerol in a 1:1 ratio with the E. coli strains containing pSPYNE-A and pSPYCE-B. Quickly freeze in liquid nitrogen for 5 minutes, then store in a -80°C freezer for long-term preservation.
List of Reagents and Equipment Used:
a.TIANprep Mini Plasmid Kit (TIANGEN BIOTECH, DP103)
b.Spin Columns CP3
c.Collection Tubes
d.Balance liquid BL
e.Solution P1 containing RNase A
f.Solution P2
g.Solution P3
h.Washing liquid PW with added anhydrous ethanol
i.Deionized water (ddH2O)
j.Tabletop microcentrifuge
k.Pipettes and tips
9.1 Equilibration of Spin Columns CP3: Add 500 μl of Buffer BL to Spin Columns CP3 (place Spin Columns into Collection Tubes). Centrifuge at 12,000 rpm (~13,400×g) for 1 minute. Discard the flow-through in the collection tube and place the Spin Columns CP3 back into the Collection Tubes.
9.2 Bacterial Cell Harvest: Take 2 ml of overnight cultured E. coli liquid containing pSPYNE-A and pSPYCE-B, centrifuge at 12,000 rpm (~13,400×g) for 1 minute in a conventional tabletop centrifuge. Remove as much supernatant as possible (if there is a large volume of bacterial liquid, centrifugation can be repeated to collect the bacterial pellet in one tube).
9.3 Resuspension of Bacterial Pellet: Add 250 μl of Buffer P1 (please check if RNase A has been added) to the centrifuge tube containing the bacterial pellet, resuspend the bacterial pellet thoroughly using a pipette or vortex mixer.
Note: If there are any unmixed bacterial clumps, it will affect lysis, leading to lower yield and purity of the extracted DNA.
9.4 Lysis and Neutralization: Add 250 μl of Buffer P2 to the centrifuge tube, gently invert the tube 6-8 times to ensure complete lysis of the bacteria.
Note: Mix gently and avoid vigorous vortexing to prevent shearing of genomic DNA. The lysate should become slightly viscous and clear. The lysis reaction should not proceed for more than 5 minutes to avoid plasmid damage. If the lysate is not clear, reduce the amount of bacterial pellet due to incomplete lysis.
9.5 Neutralization and Precipitation: Add 350 μl of Buffer P3 to the centrifuge tube, mix immediately and thoroughly by inverting the tube 6-8 times. A white flocculent precipitate should form. Centrifuge at 12,000 rpm (~13,400×g) for 10 minutes.
9.6 DNA Binding: Carefully transfer the supernatant from the previous step to the Spin Columns CP3 (place Spin Columns CP3 into Collection Tubes) using a pipette without disturbing the precipitate. Centrifuge at 12,000 rpm (~13,400×g) for 60 seconds. Discard the flow-through in the collection tube and place the Spin Columns CP3 back into the Collection Tubes.
9.7 Washing of Spin Columns CP3: Add 600 μl of Buffer PW (please check if anhydrous ethanol has been added) to the Spin Columns CP3, centrifuge at 12,000 rpm (~13,400×g) for 30-60 seconds. Discard the flow-through in the collection tube and place the Spin Columns CP3 back into the Collection Tubes.
9.8 Repeat Washing: Repeat the operation in step 9.7.
9.9 Drying of Spin Columns CP3: Place the Spin Columns CP3 into the Collection Tubes, centrifuge at 12,000 rpm (~13,400×g) for 2 minutes, then let stand at room temperature for 10 minutes to completely dry the adsorbent material and remove any residual wash buffer.
9.10 Elution of Plasmid DNA: Place the Spin Columns CP3 in a clean 1.5 ml microcentrifuge tube, add 30 μl of ddH2O to the center of the Spin Column CP3 membrane, let stand at room temperature for 2 minutes, then centrifuge at 12,000 rpm (~13,400×g) for 2 minutes to collect the plasmid solution into the tube. The final plasmid DNA of pSPYNE-A and pSPYCE-B can be stored at -20°C after measuring the concentration.
References:
For detailed procedures on plasmid extraction using the TIANprep Mini Plasmid Kit, refer to the manufacturer's protocol:
TIANGEN BIOTECH. TIANpure Mini Plasmid Kit Handbook.
10. Agrobacterium Transformation of Recombinant Plasmids pSPYNE-A and pSPYCE-B
List of Reagents and Equipment Used:
a.pSPYNE-A and pSPYCE-B recombinant plasmids
b.Agrobacterium tumefaciens competent GV3101 cells
c.Liquid Luria-Bertani (LB) medium
d.Kanamycin (Kan) at a concentration of 50 μg/ml
e.Rifampicin (Rif) at a concentration of 50 μg/ml
f.Solid LB agar plates
g.Constant temperature incubator
h.Water bath
i.Shaking incubator
j.Microcentrifuge tubes
10.1 Preparation of Agrobacterium Transformation Mixture: Withdraw 5 μl of the final carrier plasmids pSPYNE-A and pSPYCE-B (approximately 1-2 μg) and add them to 100 μl of Agrobacterium tumefaciens competent GV3101 cells to mix uniformly.
10.2 Cold and Heat Shock Treatment: Place the mixture on ice for 30 minutes, then quickly immerse in liquid nitrogen for 5 minutes, followed by a 5-minute incubation at 37°C in a water bath, and then immediately place on ice for 2 minutes.
10.3 Recovery in Liquid Media: Add 800 μl of liquid Luria-Bertani (LB) medium to the mixture and incubate at 28°C with shaking at 200 rpm for 4 hours.
10.4 Spreading on Solid LB Plates and Incubation: Withdraw 200 μl of the bacterial culture and spread it onto solid LB plates containing 50 μg/ml Kanamycin and 50 μg/ml Rifampicin. Incubate at 28°C in a constant temperature incubator for 48 hours.
References:
For detailed procedures on Agrobacterium transformation, refer to:
Hoeffler, U. (2017). Microbiological Methods: Agrobacterium-Mediated Transformation. In: J. W. Pollard & M. N. Malim (Eds.), Aids to Molecular Biology: Volume 1. Oxford: IRL Press.
Sambrook, J., & Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual (3rd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
11. Cultivation and Management of Nicotiana benthamiana
List of Reagents and Equipment Used:
a.Ethanol (50-60%)
b.Sodium hypochlorite solution (10%)
c.Sterile water
d.Moist sterile filter paper
e.Petri dishes
f.Plastic wrap or lids
g.Sterile soil or growth medium
h.Seed trays or pots
i.Greenhouse or growth chamber
j.Moisture meter or similar device for checking soil humidity
11.1 Disinfection: Immerse the seeds in 50-60% ethanol for approximately 1 minute, followed by rinsing with sterile water three times. Subsequently, soak the seeds in a 10% sodium hypochlorite solution for about 10 minutes, and rinse with sterile water three times.
11.2 Germination: Place the disinfected seeds on moist sterile filter paper within a petri dish, seal with plastic wrap or a plastic lid. Incubate the petri dish at 28°C in the dark to promote seed germination, which typically takes 3 days.
11.3 Soil Preparation: Fill seed trays or pots with sterile soil or a specialized growth medium, ensuring the soil is moist and the pH is appropriate (usually 6.0-6.5).
11.4 Sowing: Sow the germinated seeds on the soil surface, lightly covering them with a thin layer of soil.
11.5 Greenhouse or Growth Chamber Cultivation: Place the sown seed trays in a greenhouse or growth chamber, providing ample light under a photoperiod of 14 hours light/10 hours darkness, at a temperature of 25°C and a relative humidity of 70%. Cultivate for approximately 4-5 weeks. Regularly check the soil moisture to keep the soil moist but not waterlogged.
11.6 Standard Practice: When the plants have developed 4-6 true leaves, they are ready for Agrobacterium-mediated transformation.
References:
For detailed information on the cultivation and management of Nicotiana benthamiana, refer to:
Goodin, M. M., et al. (2008). Nicotiana benthamiana: Its History, Biology, and Role as a Model Organism for Plant Biology Research. In: Plant Biotechnology: Current and Future Uses of Genetically Modified Crops. Wallingford, UK: CAB International.
Odell, J. T., et al. (2004). Plant Transformation: A Practical Approach. Oxford: Oxford University Press.
12. Transient Expression of Fungal Effector Protein A and Plant Receptor Protein B in Nicotiana benthamiana Leaf Tissue
Note: Preliminary work also requires the transformation of empty vectors pSPYNE and pSPYCE into Agrobacterium, following the same transformation steps as pSPYNE-A and pSPYCE-B.
List of Reagents and Equipment Used:
a.pSPYNE-A and pSPYCE-B recombinant plasmids
b.Agrobacterium tumefaciens GV3101 competent cells
c.Kanamycin (Kan) at a concentration of 50 μg/ml
d.Rifampicin (Rif) at a concentration of 50 μg/ml
e.Liquid LB medium
f.Acetosyringone
g.Infiltration buffer X (10 mM MgCl2, 10 mM MES, 150 μM acetosyringone, pH = 5.6)
h.Nicotiana benthamiana plants
i.1 ml needle
j.Marker for marking leaves
k.Greenhouse or growth chamber
l.Camera for documentation
12.1 Inoculation of Agrobacterium Single Colonies:Select single colonies of Agrobacterium containing the final vectors pSPYNE-A and pSPYCE-B and inoculate them into 5 ml of LB medium containing 50 μg/ml Kanamycin and 50 μg/ml Rifampicin. Cultivate at 28°C with 200 rpm shaking for 2 days. (Freshly transformed Agrobacterium single colonies can be cultured overnight in 3 ml of medium until day 1.)
12.2 Liquid Culture and Expansion: Transfer 1 ml of the cultured Agrobacterium liquid to 20 ml of liquid LB medium containing 50 μg/ml Kanamycin and 50 μg/ml Rifampicin for expanded culture. This LB medium also contains 15 μM acetosyringone. Cultivate at 28°C with 200 rpm shaking until the Agrobacterium reaches the logarithmic growth phase (OD600 = 0.5-0.6).
12.3 Collection and Resuspension of Bacterial Cells:Centrifuge at 5,000 rpm for 10 minutes at room temperature to collect the bacterial cells. Resuspend the Agrobacterium cells in infiltration buffer X (containing 10 mM MgCl2, 10 mM MES, 150 μM acetosyringone, pH = 5.6) to an OD600 of 1.0. Allow the cells to stand at room temperature for 2 hours.
12.4 Mixture of Bacterial Cultures and Infiltration: Mix equal volumes of the two bacterial cultures containing pSPYNE-A and pSPYCE-B. Use a 1 ml needle to gently make a small incision on the abaxial side of a Nicotiana benthamiana leaf (be careful not to pierce through). Then, use a needle without a syringe toabsorb the bacterial suspension and inject it into the leaf through the wound. Mark the area on the leaf with a marker.
Note: The injection combinations are pSPYNE-A+pSPYCE-B, pSPYNE-A+pSPYCE, pSPYNE+pSPYCE, and pSPYNE+pSPYCE-B.
12.5 Cultivation and Phenotypic Observation: Cultivate the injected plants in the dark at approximately 25°C for 5 days, then observe for phenotypes in the areas of the tobacco leaves infiltrated with Agrobacterium, and save photographs using a camera.
Supplementary: Solution Formulas
0.5 M MES (pH 5.6): Weigh out 9.75 g of anhydrous MES and dissolve in deionized water. Adjust the pH to 5.6 with NaOH, bring to a final volume of 100 ml, filter sterilize, and store at 4°C.
150 mM Acetosyringone: Weigh out 2.943g of acetosyringone and dissolve in 5 ml of DMSO (dimethyl sulfoxide). Add deionized water to bring to a final volume of 10 ml, filter sterilize, and store at -20°C.
Note:1 mM=1000 μM
References:
For detailed procedures on Agrobacterium-mediated transient expression in Nicotiana benthamiana, refer to:
Keshavareddy, G., Kumar, A.R.V., & Ramu, V.S. (2018). Methods of Plant TransformationA Review. International Journal of Current Microbiology and Applied Sciences, 7(7),2656-2668.
Yuan, M. and Xu, C. Y. (2018). BiFC assay for detecting protein-protein interaction in
tobacco leaves. Bio-101: e1010133. Doi: 10.21769/BioProtoc.1010133. (in Chinese)
Chen, X. S., Li, T. T., Zhou, S. L. and Zhao, Y. (2018). Transient expression of exogenous
protein in tobacco leaves. Bio-101 e1010127. Doi: 10.21769/BioProtoc.1010127. (in Chinese)
- Wang, S, Huang, Z, Liu, Y, Shao, S, Li, L and Ma, M(2024). Application of the Nicotiana Allergic Necrosis Assay for the Validation of Protein-Protein Interactions between Fungal Effectors and Plant Receptor Kinases. Bio-protocol Preprint. bio-protocol.org/prep2729.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
