Advanced Search
Lentiviral transductions and engineering of stable cell lines
Last updated date: Oct 19, 2023 Views: 720 Forks: 0
Lentiviral Packaging:Transfection of Adherent 293T Cells with Calcium Chloride
Materials Needed:
- 293T growth medium (DMEM 4.5 g/L glucose, 10% FBS, 1% Penicillin/Streptomycin, 1% Sodium Pyruvate)
- 293T transfection medium (DMEM 4.5 g/L glucose, 10% FBS, 1% Sodium Pyruvate)
- Lenti-X 293Tcells
- Tissue culture vessel
- PBS (no calcium, no magnesium)
- 0.05% Trypsin-EDTA
- Lentivirus packaging plasmids
- Lentiviral transfer plasmid with geneof interest
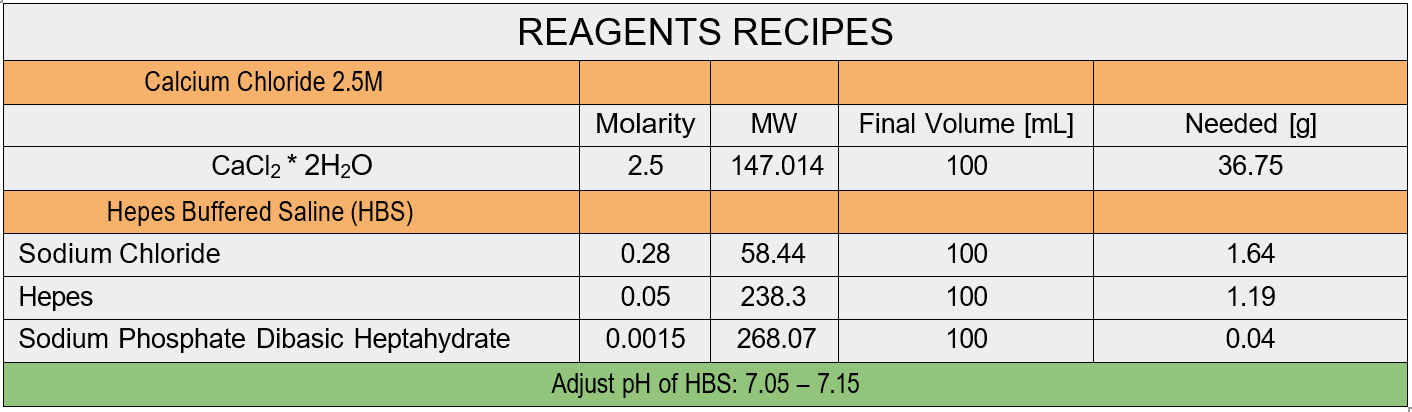
- 2.5 M Calcium Chloride
- 2X HepesBuffered Saline (pH 7.1)
Protocol:
Day 1. Plate cells approximately 18-24h before transfection:
- Aspirate the medium from a healthyand fast-growing culture of 293T cells
- Wash gently with PBS
- Detach the cells with 0.05% Trypsin-EDTA and count them
- Seed the 293T cells in growth medium in the desired vessel. Cells should be 65-75% confluent on the following day. As a rule of thumb, seed 80,000 cells/cm2.
- Culture the cells overnight
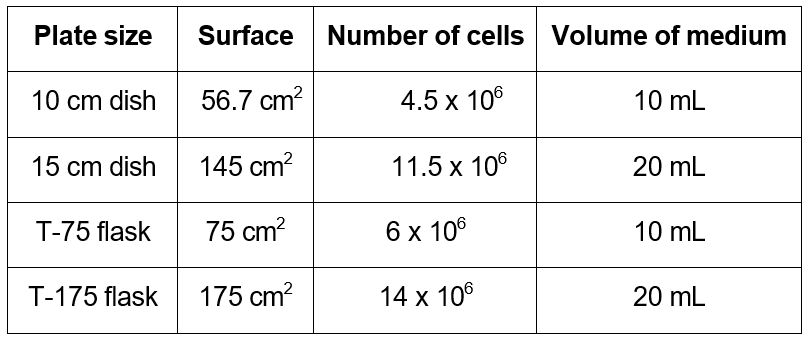
Day 2. Production of DNA complexes and transfection:
1. Before transfecting, make sure that your cells are 65-75% confluent (rough estimate picture below)
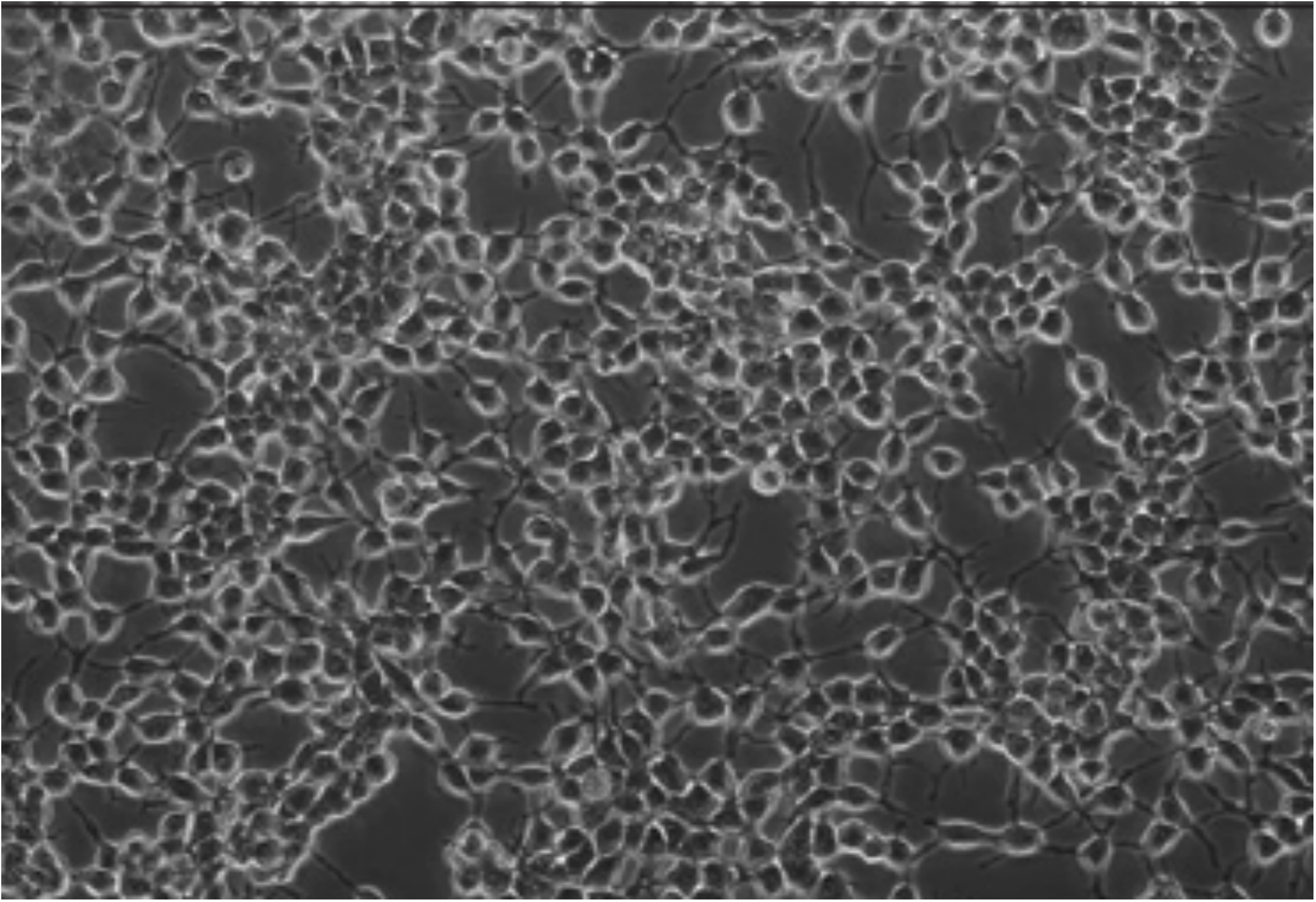
2. 2-4 hours prior to transfection, gently change the medium to fresh transfection medium
3. Prepare two 15 mL conical tubesas follows
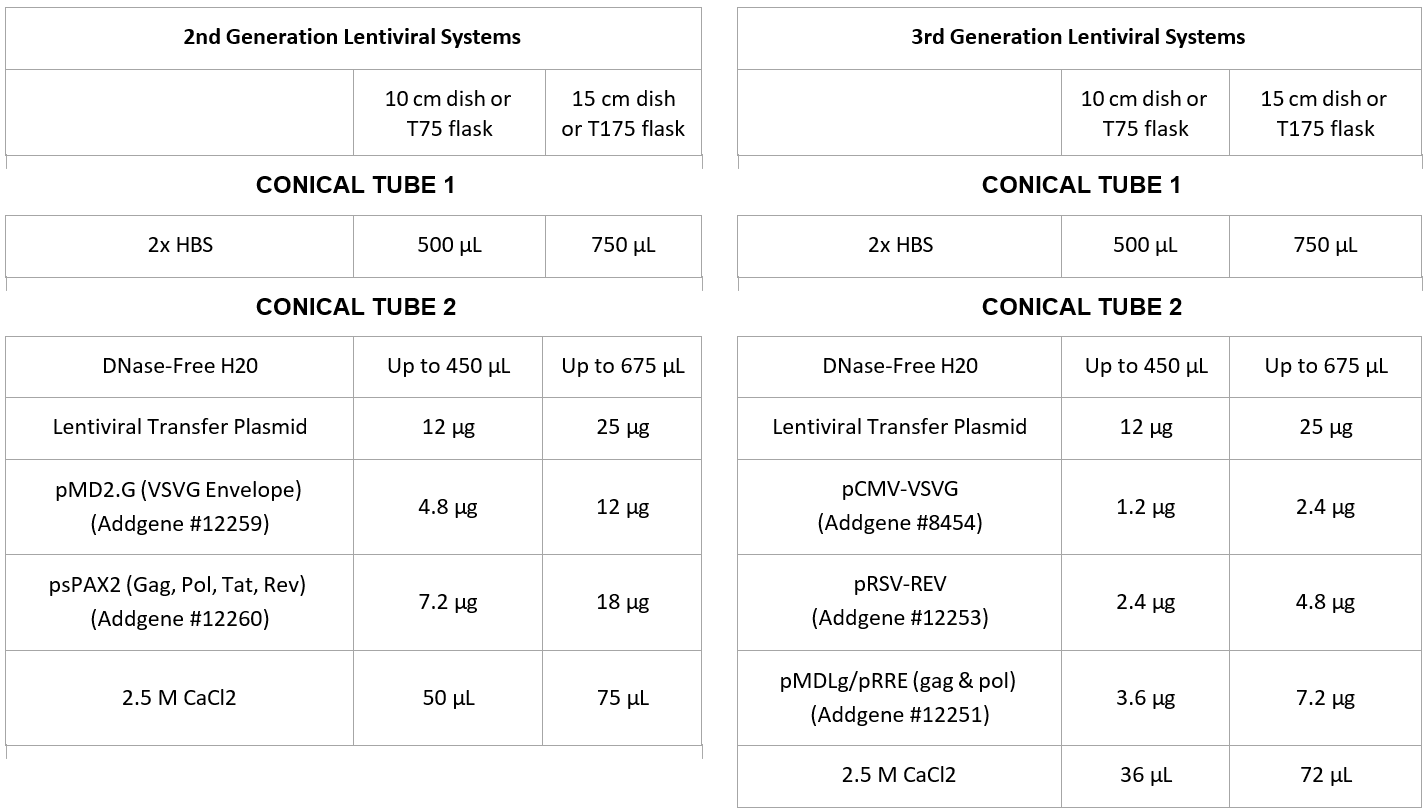
4. Gently mix Tube
5. While vortexing Tube 1, add the contents of Tube 2, drop-wise, into Tube 1
6. Incubate at RT for 30 mins, vortexing every 10 minutes for 30 seconds
7. Add the solution dropwise homogeneously to the 293T cells
8. Gently rock the culture vessel back-and-forth and from side-to-side to evenly distribute Reagent: DNA complexes
Day 3. Medium change:
- After 16 hours, change the medium with fresh growth medium. Use half the original volume to obtaina more concentrated supernatant
- Incubate at 37°C in 5% CO2 for 24h
Day 4. Collection:
- Collect the supernatant without disturbing the cells and store it in a new, sterile conical tube
- Optional: add fresh growth medium to the cell culture and collect the viral supernatant again after 24h
- Centrifuge the collected supernatant at 750 x g for 5 minutes to pellet debris.
- Filter using a 0.45 um PES or SFCA syringe filter into a new conical tube.
- The supernatant is ready for concentration and/or storage.
VIRUS CONCENTRATION BY ULTRACENTRIFUGATION
Handling virus-containing supernatant requires proper decontamination of the disposable supplies (pipets, tips, syringes, filters, tubes etc.) before trashing. Please discard tips and filters in the bottle with bleach and rinse all syringes and serological pipettes with bleach before discarding.
Materials Needed:
- PBS
- 60 mL syringes
- Syringe filter (0.45µL PES)
- Ultracentrifuge tubes
- 70% Isopropanol
- Virus vials
- Eppendorf tubes
- Turn on the ultracentrifuge, place the rotor inside (without the buckets), set the temperature to 4°C and activate the vacuum pump to decrease the temperature.
- Spin down the virus-containing supernatant at 2000 RPM for10 min to pellet cell debris.
- Filter the supernatant with 0.45 µm PES filters (if PES is not available, SFCA is a valid alternative. Please do not use CN or Nylon filters, these materials bind proteins)
- Sterilize the number of buckets and ultracentrifuge tubes necessary to hold all your virus (consider 36 mL per tube) by filling them with 70% isopropanol for 25 minutes OR byleaving them in the hood with UV on for 20 min.
- If using isopropanol, remove it (can be reused up to 3 times) and let tubes, buckets, and lids air dry completely.
- Insert ultracentrifuge tube in the bucket and equally splitthe filtered viral supernatant into each tube.
- Weight the buckets to ensure they are of the same weight (for centrifuge balancing purposes). If not, add PBS to the buckets to balance.
- Attach buckets to the rotor, making sure they are in the right position (numbers matching in lids, buckets, and rotor) and properly balanced.
- Centrifuge at 120,000 RCF for 2 h in ultracentrifuge using a swinging-bucket rotor.
- From now on, all procedures should be performed on ice.
- Collect the buckets from the ultracentrifuge and bring them under the hood. Take out the ultracentrifuge tubes from the buckets and remove the supernatant by tilting the tube while aspirating with the vacuum.
- Leave the emptied ultracentrifuge tube upside down on absorbent paper for a couple of minutes to absorb the latest drops of liquid.
- The presence of a pellet is not a guarantee of success. In fact, what makes a pellet visible is cell debris. The virus pellet is invisible.
- Resuspend the pellet in PBS (the amount is your choice)
- Pool the resuspended virus in a 1.7 mL Eppendorf tube and spin it down at full speed for 1 min at 4°C in the benchtop centrifuge
- Aliquot the supernatant in virusvials for freezing
TRANSDUCTION OF CELL LINES USING VIRUSES WITHOUT ANTIBIOTIC RESISTANCE GENES
Materials Required:
- 6-well plate
- Culture medium
- Protamine sulfate 4 mg/mL
- Virus of interest
- Fluorescence microscope
Day 1
- Plate 200,000 cells per well in 3 wells of a 6-well plate.
Day 2
- Change the medium (2 mL fresh culture medium) and add 6 µL of protamine sulfate 4 mg/mL.
- Note: protamine sulfate increases the cell membrane permeability, facilitating the infection.
- Protamine sulfateis a substitute for polybrene (hexadimethrine bromide)
- Add your virus of interest to the different wells, using MOIs of 2, 5, and 10.
- For some easy-to-transduce cell lines, you can use an unconcentrated virus-containing medium.
Day 3
- 12-24 hours later (optimize for your needs), change the medium of the transduced cells to remove the toxic polybrene/protamine sulfate/ excess virus particles
- Check under a fluorescence microscope the expression of color genes (GFP takes approximately 24-36 hours to be expressed, and mCherry 48 hours).
- Subculture the well with the best transduction efficiency and healthier cells. OPTIONAL: Based on transduction efficiency and experimental plan 7 & 8 might be necessary
- After one passage, sort the cells for GFP or mCherry.
- Sort the cells using FACS to ensureall the future sub cultured cells are engineered ones.
- Expand the transduced cells,verify gene expression and bank. If the vector contains other genes (luciferases or secreted molecules), confirm that these genes are also expressed and functional.
Note: If you do not sort your cells, you may find that the wild-type cells will outgrow the transduced cells in some cases.
TRANSDUCTION OF CELL LINES USING VIRUSES WITH ANTIBIOTIC RESISTANCE GENES
Prior to starting the transduction experiment or in concert, please perform a Dose titration with the antibiotic or selection agent being used to determine the optimal concentration of antibiotic required.
Materials Required:
- 6-well plate
- Culture medium
- Protamine sulfate 4 mg/mL
- Virus of interest
- Fluorescence microscope
- Selection agent/ antibiotic
- Plate 200,000 cells per well in 3 wells of a 6-well plate.
- The next day, change medium (2 mL fresh culture medium) and add 6 µL of protamine sulfate 4 mg/mL.
- Note: protamine sulfate increases the cell membrane permeability, facilitating the infection.
- Note: protamine sulfate is a substitute for polybrene
- Add your virus of interest to the different wells, using MOIs of 2, 5, and 10.
- Approximately 16-24 hours later, change the medium of the transduced cells.
- Check under fluorescence microscope the expression of color genes (GFP takes approximately 24-36 hours to be expressed, and mCherry 48 hours).
- Subculture the well with the best transduction efficiency and healthier cells. Once you have enough cells, in the morning plate 200,000 cells per well in a 6-well plate (create a plate with transduced cells and another with wild type to use as a control).
- In the evening, add the selection antibiotic at concentrations previously determined from the titration experiment.
- The next day, check for cell death in both transduced and wild type cells. Subsequent treatment might be essential based on efficiency.
- Expand the selected cells and bank. If the vector contains other genes (luciferases or secreted molecules), confirm that these genes are also expressed and functional.
Note: The optimal antibiotic concentration to use is the lowest concentration that kills 100% of the wild type cells in 48 hours from the start of the puromycin selection.
Note: puromycin is commonly used as a selection marker; however, it is very immunogenic and can affect in vivo experiments. For this reason, it is NOT recommended to use lentiviruses with a puromycin resistance gene to engineer murine tumor lines.
- Chen, K and Shah, K(2023). Lentiviral transductions and engineering of stable cell lines. Bio-protocol Preprint. bio-protocol.org/prep2473.
- Chen, K., Reinshagen, C., Van Schaik, T. A., Rossignoli, F., Borges, P., Mendonca, N. C., Abdi, R., Simon, B., Reardon, D. A., Wakimoto, H. and Shah, K.(2023). Bifunctional cancer cell–based vaccine concomitantly drives direct tumor killing and antitumor immunity. Science Translational Medicine 15(677). DOI: 10.1126/scitranslmed.abo4778
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
