Advanced Search
Detection and Analysis of human ALDH2 by quantitative RT-PCR
Last updated date: Oct 7, 2023 Views: 352 Forks: 0
Yao-Hui Gao1, Guo-Qiang Chen2*, Li-Shun Wang3,4*
1. Department of Pathology, Shanghai Tenth People's Hospital, Tongji University School of Medicine, 200072 Shanghai, China.
2. State Key Laboratory of Oncogenes and Related Genes, and Chinese Academy of Medical Sciences Research Unit (NO.2019RU043), Renji Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China
3. Center for Traditional Chinese Medicine and Gut Microbiota, Minhang Hospital, Fudan University, 201199 Shanghai, China;
4. Institute of Fudan-Minhang Academic Health System, Minhang Hospital, Fudan University, 201199 Shanghai, China.
*Correspondence:
Li-Shun Wang (lishunwang@fudan.edu.cn);
Guo-Qiang Chen (chengq@shsmu.edu.cn);
Abstract
Acetaldehyde dehydrogenase (ALDH) is a member of the oxidase family responsible for detoxifying aldehydes. Acetaldehyde dehydrogenase 2(ALDH2)is highly expressed in liver, heart, brain, and kidney. It is known for its key role in acetaldehyde metabolism. ALDH2 also plays a key role in the metabolism of endogenous aldehydes produced during lipid peroxidation stress, such as 4-hydroxy-2-Nonenal (4-HNE), and is therefore considered to be a key enzyme in protecting the heart from oxidative stress. ALDH2 dysfunction may be associated with the occurrence and development of a variety of diseases, such as cardiovascular disease, liver disease, and cancer. In addition, ALDH2 has been found to play an important role in chemotherapy drug sensitivity in cancer. To date, activators and inhibitors of ALDH2 have been used in animal models and preclinical studies. In view of the importance of ALDH2 and potential clinical application, we established a quantitative RT-PCR-based protocol to detect the transcriptional expression of ALDH2.
Keywords: ALDH2, mRNA, Primer, Quantitative RT-PCR
Background
The human acetaldehyde dehydrogenase (ALDH) gene family consists of 19 putative members (Zakhari and Li, 2007). Aldehyde dehydrogenase 2 (ALDH2) is a key enzyme for the detoxification of acetaldehyde and endogenous lipid aldehydes such as 4-hydroxy-2-nonenal (4-HNE), which are generated from lipid peroxidation (X. Chen et al., 2021). ALDH2 is composed of 13 exons on chromosome 12 (12q24) and localizes to mitochondria (Matsumoto, 2016). The detection of ALDH2 gene polymorphisms identified 535 coding single nucleotide polymorphisms (SNPs). Among them, the rs671 SNP is present in approximately 30–50% of the East Asian population (J. Zhang et al., 2023). The rs671 SNP causes a substantial reduction in dehydrogenase activity, leads to Asian flushing syndrome, which is characterized by skin flushing after drinking alcohol among East Asian people (Zambelli et al., 2014).
The role of ALDH2 rs671 SNP in the development of different types of cardiovascular disease (CVD) has been widely explored (J. Zhang et al., 2023). ALDH2 rs671 SNP may accelerate the occurrence and development of CVD by regulating the biological processes such as apoptosis, necrosis and autophagy of cardiomyocytes (Ma et al., 2011; Sun et al., 2014; Fang et al., 2018). However, ALDH2 rs671 SNP has been shown to have a protective effect against cardiac dysfunction and aortic aneurysm or dissection (Y. Zhang et al., 2014; K. Yang et al., 2020). ALDH2 expression or dysfunction is also associated with liver disease, such as liver inflammation and fibrosis, non-alcoholic fatty liver disease, liver fibrosis, and liver cancer (Wang et al., 2020). Notably, an independent hepatocellular carcinoma (HCC) cohort study shows that ALDH2 expression is inversely associated with the development of malignant HCC (Hou et al., 2017). In terms of cancer therapy, our study shows that the expression of ALDH2 correlates with the sensitivity of anthracycline chemotherapeutic agents (Gao et al., 2017). These results suggests that ALDH2 has emerged as a promising therapeutic target for cancer therapy.
The therapeutic potential of ALDH2 activator Alda-1 in CVD has been demonstrated in preclinical studies (C. H. Chen et al., 2014). Alda-1 doubles the catalytic activity of ALDH2 and increases the ALDH2 enzyme activity of individuals with ALDH2 * 2/2 genotype by 11-fold (C. H. Chen et al., 2008; C. H. Chen et al., 2014). It is reported that Alda-1 reduces the infarct area of myocardial ischemia in rats by 60% by removing cytotoxic aldehydes (C. H. Chen et al., 2008). In addition, the cardioprotective capacity of Alda-1 also involves mitophagy and signaling pathways such as SIRT1, TGF β and Wnt/β-catenin (Gu et al., 2013; Yuan et al., 2019; Zhao et al., 2015). Daidzin has been widely used as an ALDH2 inhibitor which binds in a hydrophobic pocket between the catalytic and NAD+ -binding domains of ALDH2 (Lowe et al., 2008). Daidzin inhibits aortic aneurysm formation in a model of atherosclerosis and increases oxidative low-density lipoprotein-mediated apoptosis (K. Yang et al., 2020; M. Y. Yang et al., 2018).
Although great progress has been made in the functional study of ALDH2 in recent years, the mechanism of ALDH2 regulation has not been fully elucidated. We established a RT-PCR-based protocol to detect the transcriptional expression of human ALDH2, which provides a basis for studying the molecular regulation mechanism of ALDH2.
Materials and Reagents
- Standard pipette tips with a volume capacity of 10 µl, 200 µl, and 1 ml (Eppendorf, Germany).
- 1.5ml centrifuge Tubes ( catalog number: 0030120086,Eppendorf, Germany)
- TRIzol Lysis Reagent (catalog number:15596018,Invitrogen, USA)
- Chloroform (Lingfeng Chemical Reagent Co., Ltd., China)
- PBS buffer (catalog number: E607008, Sangon Biotech, China)
- Isopropanol (Lingfeng Chemical Reagent Co., Ltd., China, China)
- Ethanol (Lingfeng Chemical Reagent Co., Ltd., China, China)
- DEPC H2O (catalog number: B501005, Sangon Biotech, China)
- TaKaRa RNA PCR kit (AMV) (catalog number: RR019A,TaKaRa,Japan )
- The double-stranded DNA dye SYBR Green PCR Master Mixture Reagents (catalog number: 4367659,Applied Biosystems, USA)
Equipment
- Manual Pipettes set of 2.5 µl, 20 µl, 200 µl and 1 ml (Eppendorf, Germany)
- ABI PRISM® 7500HT ( Applied Biosystems,USA)
- NanoDrop spectrophotometer(Thermo Scientific,USA)
Procedure
A. Primer design
1. Get the mRNA sequence of ALDH2 from NCBI (National Center for Biotechnology Information) Web Pages (https://www.ncbi.nlm.nih.gov/gene/).
2. Use the above mRNA sequence to design PCR primers using Primer Premier Software or NCBI primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The following specific primers are designed for detection of ALDH2 (Table 1).
Table 1. The specific primer for detection of ALDH2.

Note: F: Forward ; R: Reverse
3. The specificity of primers was confirmed by NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/). As shown in Figure 1, the designed primers are specific for human ALDH2.
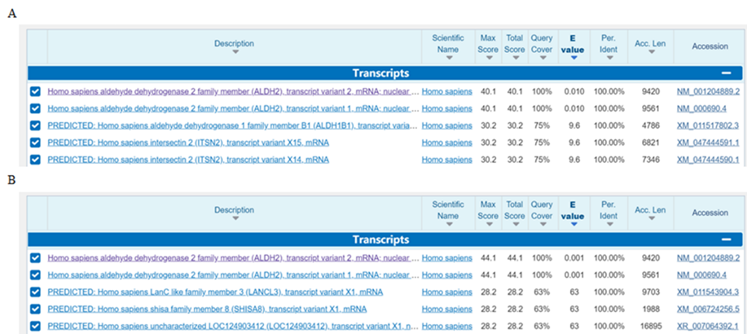
Figure 1. The BLAST results of ALDH2 primers. (A) Forward primer. (B) Reverse primer.
B. Total RNA isolation and reverse transcription
- Collect cultured cells and remove the culture media.
- Wash the cells three times with cold PBS at 4 °C.
- Collect the cell pellet by centrifugation at 800 g for 5 min at 4 °C.
- Add 1 ml of TRIzol Lysis Reagent and vortex.
- Incubate at room temperature for 5 minutes.
- Add 200μl of chloroform, turn it upside down vigorously for 15 seconds, and incubate at v for 10 minutes.
- Centrifuge at 12,000 g for 15 minutes at 4 °C, and transfer the supernatant to a fresh tube.
- Add equal volume of isopropanol to the supernatant, mix it upside down several times vigorously, and incubate it at room temperature for 10 minutes.
- Centrifuge at 12,000 g for 15 minutes at 4 °C and remove the supernatant.
- Add 1ml of 70% ethanol to wash pellet, centrifuge at 13,000 g for 5 min. Carefully discard the supernatant.
- Briefly dry the RNA pellet. Dissolve the RNA in 50μl RNase-free water.
- Measure RNA concentration with a NanoDrop spectrophotometer. A260/ A280 should be between 1.9 and 2.2.
- Reverse transcription was performed with TaKaRa RNA PCR kit (AMV) following the manufacturer’s instructions.
C. Quantitative RT-PCR
- The double-stranded DNA dye SYBR Green PCR Master Mixture Reagents was used for Quantitative RT-PCR.
- The assay was carried out in a 10 μl reaction mixture containing:

3. Set up the experiment and the following PCR program on ABI 7500 Fast Real-Time PCR System.
- 50°C for 2 min, 1 cycle
- 95°C for 10 min, 1 cycle
- 40 cycles at 95 °C for 15 s ,60 °C for 30 s, and 72 °C for 30 s, followed by a melting curve analysis program.
- 72°C for 10 min, 1 cycle.
4. Analyze the result with the analyze software. Check if there is any bimodal or other abnormal dissociation curve.
5. For comparison of the relative transcript expression between the different groups, we used the 2-ΔΔCq method. Actin served as a control and the primers for Actin has been described previously (Gao et al., 2013). The folds of changes were shown as means±s.d. in three independent experiments with each triplicate.
Acknowledgments
This protocol was adapted from the previously published papers (Gao et al., 2017). The protocol was tested and optimized by different researchers in Key Laboratory of Cell Differentiation and Apoptosis of National Ministry of Education, Shanghai Jiaotong University School of Medicine. The authors have no conflicts of interest or competing interests to declare.
References
Chen, C. H., Budas, G. R., Churchill, E. N., Disatnik, M. H., Hurley, T. D. and Mochly-Rosen, D. (2008). Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 321(5895): 1493-1495. https://www.ncbi.nlm.nih.gov/pubmed/18787169
Chen, C. H., Ferreira, J. C., Gross, E. R. and Mochly-Rosen, D. (2014). Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev 94(1): 1-34. https://www.ncbi.nlm.nih.gov/pubmed/24382882
Chen, X., Li, X., Xu, X., Li, L., Liang, N., Zhang, L., Lv, J., Wu, Y. C. and Yin, H. (2021). Ferroptosis and cardiovascular disease: role of free radical-induced lipid peroxidation. Free Radic Res 55(4): 405-415. https://www.ncbi.nlm.nih.gov/pubmed/33455488
Fang, T., Cao, R., Wang, W., Ye, H., Shen, L., Li, Z., Hu, J. and Gao, Q. (2018). Alterations in necroptosis during ALDH2‑mediated protection against high glucose‑induced H9c2 cardiac cell injury. Mol Med Rep 18(3): 2807-2815. https://www.ncbi.nlm.nih.gov/pubmed/30015964
Gao, Y. H., Li, C. X., Shen, S. M., Li, H., Chen, G. Q., Wei, Q. and Wang, L. S. (2013). Hypoxia-inducible factor 1alpha mediates the down-regulation of superoxide dismutase 2 in von Hippel-Lindau deficient renal clear cell carcinoma. Biochem Biophys Res Commun 435(1): 46-51. https://www.ncbi.nlm.nih.gov/pubmed/23611775
Gao, Y. H., Wu, Z. X., Xie, L. Q., Li, C. X., Mao, Y. Q., Duan, Y. T., Han, B., Han, S. F., Yu, Y., Lu, H. J., Yang, P. Y., Xu, T. R., Xia, J. L., Chen, G. Q. and Wang, L. S. (2017). VHL deficiency augments anthracycline sensitivity of clear cell renal cell carcinomas by down-regulating ALDH2. Nat Commun 8: 15337. https://www.ncbi.nlm.nih.gov/pubmed/28643803
Gu, C., Xing, Y., Jiang, L., Chen, M., Xu, M., Yin, Y., Li, C., Yang, Z., Yu, L. and Ma, H. (2013). Impaired cardiac SIRT1 activity by carbonyl stress contributes to aging-related ischemic intolerance. PLoS One 8(9): e74050. https://www.ncbi.nlm.nih.gov/pubmed/24040162
Hou, G., Chen, L., Liu, G., Li, L., Yang, Y., Yan, H. X., Zhang, H. L., Tang, J., Yang, Y. C., Lin, X., Chen, X., Luo, G. J., Zhu, Y., Tang, S., Zhang, J., Liu, H., Gu, Q., Zhao, L. H., Li, Y., Liu, L., Zhou, W. and Wang, H. (2017). Aldehyde dehydrogenase-2 (ALDH2) opposes hepatocellular carcinoma progression by regulating AMP-activated protein kinase signaling in mice. Hepatology 65(5): 1628-1644. https://www.ncbi.nlm.nih.gov/pubmed/28027570
Lowe, E. D., Gao, G. Y., Johnson, L. N. and Keung, W. M. (2008). Structure of daidzin, a naturally occurring anti-alcohol-addiction agent, in complex with human mitochondrial aldehyde dehydrogenase. J Med Chem 51(15): 4482-4487. https://www.ncbi.nlm.nih.gov/pubmed/18613661
Ma, H., Guo, R., Yu, L., Zhang, Y. and Ren, J. (2011). Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur Heart J 32(8): 1025-1038. https://www.ncbi.nlm.nih.gov/pubmed/20705694
Matsumoto, A. (2016). [Fundamental Properties of Aldehyde Dehydrogenase 2 (ALDH2) and the Importance of the ALDH2 Polymorphism]. Nihon Eiseigaku Zasshi 71(1): 55-68. https://www.ncbi.nlm.nih.gov/pubmed/26832618
Sun, A., Zou, Y., Wang, P., Xu, D., Gong, H., Wang, S., Qin, Y., Zhang, P., Chen, Y., Harada, M., Isse, T., Kawamoto, T., Fan, H., Yang, P., Akazawa, H., Nagai, T., Takano, H., Ping, P., Komuro, I. and Ge, J. (2014). Mitochondrial aldehyde dehydrogenase 2 plays protective roles in heart failure after myocardial infarction via suppression of the cytosolic JNK/p53 pathway in mice. J Am Heart Assoc 3(5): e000779. https://www.ncbi.nlm.nih.gov/pubmed/25237043
Wang, W., Wang, C., Xu, H. and Gao, Y. (2020). Aldehyde Dehydrogenase, Liver Disease and Cancer. Int J Biol Sci 16(6): 921-934. https://www.ncbi.nlm.nih.gov/pubmed/32140062
Yang, K., Ren, J., Li, X., Wang, Z., Xue, L., Cui, S., Sang, W., Xu, T., Zhang, J., Yu, J., Liu, Z., Shang, H., Pang, J., Huang, X., Chen, Y. and Xu, F. (2020). Prevention of aortic dissection and aneurysm via an ALDH2-mediated switch in vascular smooth muscle cell phenotype. Eur Heart J 41(26): 2442-2453. https://www.ncbi.nlm.nih.gov/pubmed/32428930
Yang, M. Y., Wang, Y. B., Han, B., Yang, B., Qiang, Y. W., Zhang, Y., Wang, Z., Huang, X., Liu, J., Chen, Y. D., Ren, J., Cao, F. and Xu, Y. (2018). Activation of aldehyde dehydrogenase 2 slows down the progression of atherosclerosis via attenuation of ER stress and apoptosis in smooth muscle cells. Acta Pharmacol Sin 39(1): 48-58. https://www.ncbi.nlm.nih.gov/pubmed/28858301
Yuan, Q., Cao, S., Dong, Q., Wang, Z., Xu, Y., Han, Q., Ma, J., Wei, S., Pang, J., Yang, F., Zhang, R., Liu, B., Dai, S., Xue, L., Wang, J., Xue, M., Xu, T., Zheng, W., Xu, F., Chen, Y. and Guo, P. (2019). ALDH2 Activation Inhibited Cardiac Fibroblast-to-Myofibroblast Transformation Via the TGF-beta1/Smad Signaling Pathway. J Cardiovasc Pharmacol 73(4): 248-256. https://www.ncbi.nlm.nih.gov/pubmed/30801261
Zakhari, S. and Li, T. K. (2007). Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology 46(6): 2032-2039. https://www.ncbi.nlm.nih.gov/pubmed/18046720
Zambelli, V. O., Gross, E. R., Chen, C. H., Gutierrez, V. P., Cury, Y. and Mochly-Rosen, D. (2014). Aldehyde dehydrogenase-2 regulates nociception in rodent models of acute inflammatory pain. Sci Transl Med 6(251): 251ra118. https://www.ncbi.nlm.nih.gov/pubmed/25163478
Zhang, J., Guo, Y., Zhao, X., Pang, J., Pan, C., Wang, J., Wei, S., Yu, X., Zhang, C., Chen, Y., Yin, H. and Xu, F. (2023). The role of aldehyde dehydrogenase 2 in cardiovascular disease. Nat Rev Cardiol 20(7): 495-509. https://www.ncbi.nlm.nih.gov/pubmed/36781974
Zhang, Y., Mi, S. L., Hu, N., Doser, T. A., Sun, A., Ge, J. and Ren, J. (2014). Mitochondrial aldehyde dehydrogenase 2 accentuates aging-induced cardiac remodeling and contractile dysfunction: role of AMPK, Sirt1, and mitochondrial function. Free Radic Biol Med 71: 208-220. https://www.ncbi.nlm.nih.gov/pubmed/24675227
Zhao, X., Hua, Y., Chen, H., Yang, H., Zhang, T., Huang, G., Fan, H., Tan, Z., Huang, X., Liu, B. and Zhou, Y. (2015). Aldehyde dehydrogenase-2 protects against myocardial infarction-related cardiac fibrosis through modulation of the Wnt/beta-catenin signaling pathway. Ther Clin Risk Manag 11: 1371-1381. https://www.ncbi.nlm.nih.gov/pubmed/26392772
- Gao, Y, Chen, G and Wang, L(2023). Detection and Analysis of human ALDH2 by quantitative RT-PCR. Bio-protocol Preprint. bio-protocol.org/prep2456.
- Gao, Y., Wu, Z., Xie, L., Li, C., Mao, Y., Duan, Y., Han, B., Han, S., Yu, Y., Lu, H., Yang, P., Xu, T., Xia, J., Chen, G. and Wang, L.(2017). VHL deficiency augments anthracycline sensitivity of clear cell renal cell carcinomas by down-regulating ALDH2. Nature Communications 8. DOI: 10.1038/ncomms15337
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
