Advanced Search
Pfs25 and PfMGET RNA quantification
Last updated date: Sep 29, 2023 Views: 289 Forks: 0
PfMGET-CCp4 qRT-PCR for male-female gametocyte quantification
Before you start:
> Starting material is MagNaPure extracted Total NA (usually in hard-shell 96 wells plates), use filtertips and set-up reactions in a clean PCR cabinet.
> Prepare a plate layout in Excell for your qRT-PCR plate and calculate how much mastermix you need to prepare. Always prepare for an additional +6 reactions. Plan your ivRNA standard curve dilutions in the last column (10^8 copies/ml to 10^2 copies/ml) and one water control. For the preparation of the ivRNA standard curve, see below this protocol.
qRT-PCR with the Luna Universal Probe OneStep RT-qPCR kit (NEB, order at Bioke):
> Thaw 'Luna Reaction mix 2x' and primers/probe on ice
> Mix primers and probes by gently tapping the tube, mix Luna Reaction mix by vortexing for 5 sec, spin down all reagents (short spin, 10 sec)
> Take the 'Luna RT enzyme mix 20x' out of the freezer and place directly on ice, always keep on ice! and proceed to the preparation of the mastermix immediately.
> qRT-PCR: 15 ul of mixture + 5 ul of Total NA
- >> Prepare the mastermix according to the scheme below. Below is the schedule per reaction, calculate for your total number of reactions and always prepare for an additional +6 reactions. Add in the reagents from top to bottom, starting with ddH2O and ending with the 'Luna RT enzyme mix'. After adding the 'Luna RT enzyme mix' take a 1 ml pipet and mix well by pipetting up and down.
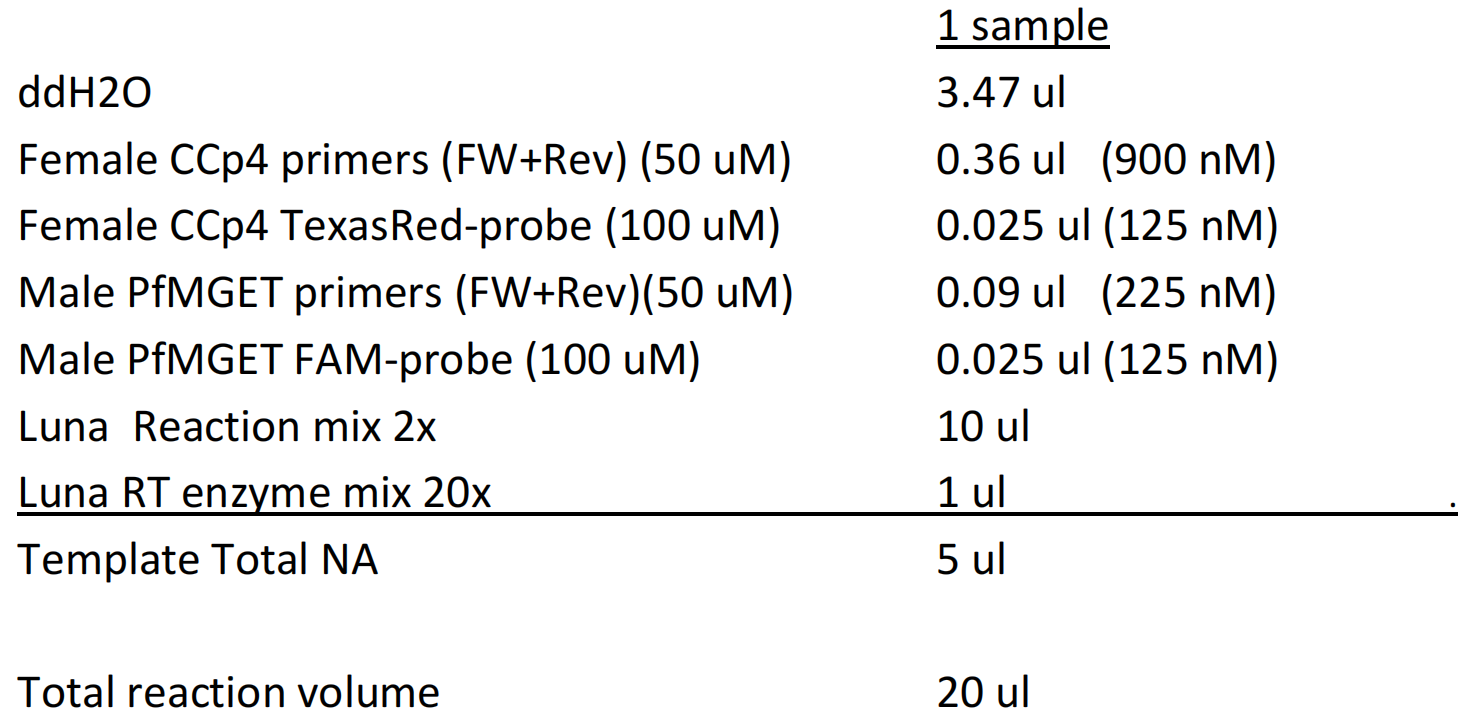
> Fill out 15 ul/well with repetitive pipet and 0.5 ml combitip in PCR cabinet. Use the BioRad hard-shell PCR 96 well plates (HSP9635)
> Add 5 ul of Total NA with multichannel and seal with plastic seal. Make sure you seal tightly, check the edges of the foil to prevent evaporation during the run.
> Spin 20 sec in plate spinner
> Run Luna One Step qRT-PCR program on BioRAD CFX96 qPCR machine: turn on PCR machine, the computer is always on > Start BioRad CFX manager > Select run type 'User defined' > Select 'select existing' > Folder Kjerstin > RTqPCR LunaOneStep > next > next > Open lid and place plate in the PCR machine (PCR program: 55℃ 15 min > 95℃ 1 min > 95℃ 10 sec, 60℃ 1 min, 44 cycles)
> Close lid > Start run: save file in folder Kjerstin
> After the run trash 96-well plate (never open plate), and turn of the PCR machine (leave computer on)
Data analysis:
> After the run the Data analysis opens automatically
> Plate setup > view/edit plate > select fluorophore FAM (PfMGET Male signal), and deselect the others
> OK
> Fill in your standard curve dilutions: first select well, fill in Sample type: standard, fill in concentration, 1,00E+08 > click load, repeat for the next dilutions > OK > apply changes: JA
> Settings > Baseline settings > Apply fluorescence drift correction
> Tick the log scale box
> Right click on amplification graph > Baseline treshold > Single threshold, User defined: set at 200 > OK
> Right click on amplification graph > Select 'Show threshold value'
> Open excel sheet and copy-paste the amplification graph and standard curve graph
> Open Quantification Data sheet > Click top left corner (all wells are selected now) > CtrlC and paste in Excell > save file on H-drive
> Go again to Plate setup > view/edit plate > select fluorophore Texas Red (CCp4 Female signal), and deselect the others > OK
> Click on top left corner in plate layout to select all wells > then click Load next to TexasRed (all the wells should now show TexasRed in them) > Fill in your standard curve dilutions: first select well, fill in Sample type: standard, fill in concentration, 1,00E+08 > click load, repeat for the next dilutions > OK > apply changes: JA
> Settings > Baseline settings > Apply fluorescence drift correction
> Right click on amplification graph > Baseline treshold > Single threshold, User defined: set at 200 > OK
> Open excel sheet and copy-paste the fluorescence curve graph and standard curve graph
> Open Quantification Data sheet > Click top left corner (all wells are selected now) > CtrlC and paste in Excell > save file on H-drive as follows >> Go to 'Bestand' (top left corner) > Select 'Opslaan als' > Double click on 'deze PC' > Select 'MMBdata H:' > Select folder 'MMB NCMLS' > Select folder 'Kjerstin' > Name your file, then click 'Opslaan'
Primers/Probes:
PfMGET Forward primer 5'- cggtccaaatataaaatcctg -3' (DST Sigma)
PfMGET Reverse primer 5'- tgtgtaacgtatgattcattttc-3' (DST Sigma)
PfMGET Probe: 5'-FAM-cagctccagcattaaaaacac-BHQ1-3' (HPLC BioLegio)
CCp4 Forward primer 5'- cacatgaatatgagaataaaattg-3' (DST Sigma)
CCp4 Reverse primer 5'- taggcgaacatgtggaaag-3' (DST Sigma)
CCp4 Probe: 5'-TexasRed-agcaacaacggtatgtgccttaaaacg-BHQ2-3' (HPLC BioLegio)
Preparation of PfMGET/CCp4 ivRNA for use as a standard curve
Order synthetic dsDNA (BaseClear), CCp4 477 bp (294669,91 ng/nmol), 450 nt ssRNA, PfMGET 527 bp (325537.47 ng/nmol), 500 nt ssRNA (in green T7-RNA promotor, in grey ssRNA, in yellow qPCR amplicon):
CCp4:

PfMGET:

> Each tube containes about 4 ug of lyophilized dsDNA. Dissolve this in 40 ul nuclease-free water which leads to a 100 ng/ul stock.
> Gel purify the dsDNA on a 1% agarose gel. Load sample over 2 wells. Cut out the correct bands and pool in one tube. Clean-up with the Qiagen Gel Purification Kit: add 500 ul QG buffer + 166 ul isopropanol. Include the extra QG wash of the standard protocol and elute in 30 ul nuclease-free water. Leave this elution step for 2 min incubation at room temp before final
spin.
> Nanodrop both samples after gel purification (in our case PfMGET 8.8 ng/ul ; CCp4 30.5 ng/ul)
> Setup the ivRNA transcription with the MEGAShortScript T7 kit (Promega), use 0.5 ml PCR tubes:

(8 ul dsDNA input is the maximum. You can calculate back what the input in the reaction was as follows:
PfMGET: 8.8 ng/ul / 325537.47 ng/nmol = 27 nM stock, 8 ul added means 11 nM in the reaction
CCp4: 30.5 ng/ul / 294669,91 ng/nmol = 104 nM stock, 8 ul added means 42 nM in the reaction)
> Run 0.5 ul of the ivRNA on a 1% agarose gel to check if transcription worked and if there is only one clear band: 0.5 ul RNA + 5 ul ddH2O + 5 ul Gel Loading buffer II (from MEGAshortscript kit). Run next to a 100 bp DNA ladder. Do a short run, let the samples run into the gel for about 2 cm. It's just to check if you have one band. (When running RNA on an agarose gel it's probably not an RNase free system and if you leave the gel running for too long you will get smearing (degradation of your
RNA). Also secondary structures might form leading to multiple bands)
> Nanodrop both samples (in our case PfMGET 10 ng/ul, CCp4 44 ng/ul)
> Setup a DNAseI treatment to get rid of your dsDNA template with the TURBO DNA-free kit DNaseI treatment (Invitrogen). Dilute samples to routine DNaseI treatment concentration. Routine = max of 2 ug of DNA in 10 ug NA/50 ul reaction:
PfMGET: 10 ul RNA + 5 ul buffer 10x + 34 ul ddH2O + 1 ul TURBO DNaseI
CCp4: 2 ul RNA + 5 ul buffer 10x + 42 ul ddH2O + 1 ul TURBO DNaseI
> Incubate for 45 min. at 37 degrees in waterbath
> Add 5 ul inactivation buffer > 5 min incubation at room temperature, flick tubes 2-3 times in between
> Spin for 1.5 min at 10.000 g > Transfer 47 ul of the supernatant to new tube
> Leftover from transcription (not DNaseI treated) + 0.5 ul RNasin > Stored at -20
> Purify the ivRNA with the RNeasy spin-column purification kit (Qiagen) according to the standard clean-up protocol. Elute in 30 ul nuclease-free water, add 0.5 ul RNasin to the eluate and store at -20
> Measure RNA concentration on a Qubit. Use the Qubit RNA HS Assay Kit which measures between 250 pg to 100 ng/ul. The actual input in the measurement should be 5-100 ng! Aim for a reading between 150-350 mg/ml, ideally around 250 mg/ml. Use 0.5 ml thin-walled Qubit Assay tubes. For CCp4 and PfMGET measure 1 ul of undiluted material. If the measurement is too close to the lower detection limit measure 2 or 3 ul, if it is too close or over to the higher detection limit dilute sample and
measure again.
> Prepare mixture: 10 ul Qubit RNA HS reagent + 1990 ul Qubit RNA HS buffer > Fill out 199 ul / 190 ul per 0.5 ml tube (and as a back-up fill out 2x 198 ul and 2x 197 ul) > Add 1 ul sample or 10 ul standard #1 and standard #2 > vortex 2 sec > Incubate RT 2 min > Measure in the Qubit on the 2nd floor. (Note: The RNA HS program is not in there, used the RNA program!).
> Calculate copy numbers using the online DNA/RNA copy number calculator: http://endmemo.com/bio/dnacopynum.php. PfMGET 500 nt ssRNA, CCp4 450 nt ssRNA, filled in the specific RNA sequence (in our case PfMGET 20.5 ng/ul which converts to 9.707*10^13 copies/ml ; CCp4 39.1 ng/ul which converts to 2.301*10^14 copies/ml)
> Dilute ivRNA samples in MagNAPure elution buffer. Mix PfMGET and CCp4 in one tube. First dilutions 10^13 to 10^9 copies/ml in Eppendorf tubes, from 10^8 copies/ml to 10^2 copies/ml in a 96-wells qPCR plate. Prepare one stock plate with multiple standard curves which can be stored at -20. For each dataset transfer one standard curve from this stock plate to a new plate which can be used for the Male-Female MPX runs (and undergo multiple freeze/thaw cycles).
- Reuling, I and Bousema, T(2023). Pfs25 and PfMGET RNA quantification. Bio-protocol Preprint. bio-protocol.org/prep2448.
- Reuling, I. J., van de Schans, L. A., Coffeng, L. E., Lanke, K., Meerstein-Kessel, L., Graumans, W., van Gemert, G., Teelen, K., Siebelink-Stoter, R., van de Vegte-Bolmer, M., de Mast, Q., van der Ven, A. J., Ivinson, K., Hermsen, C. C., de Vlas, S., Bradley, J., Collins, K. A., Ockenhouse, C. F., McCarthy, J., Sauerwein, R. W. and Bousema, T.(2018). A randomized feasibility trial comparing four antimalarial drug regimens to induce Plasmodium falciparum gametocytemia in the controlled human malaria infection model. eLife. DOI: 10.7554/eLife.31549
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
