Advanced Search
High-resolution 3D tissue imaging
Last updated date: Jul 14, 2023 Views: 1015 Forks: 0
High-resolution 3D tissue imaging protocol in mice
Lu Mu1, Xueqiang Xu2, Xuebing Yang1, Yan Zhang1, Hua Zhang1*
- State Key Laboratory of Agrobiotechnology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
- Biomedical Pioneering Innovation Center, School of Life Sciences, Peking University, Beijing 100871, China; Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China.
*Corresponding author: Hua Zhang, huazhang@cau.edu.cn
Detailed protocol
This protocol realized a whole-mount imaging system to visualize ovarian blood vessels with a single-cell resolution. This was achieved by combining a vascular endothelium–specific endogenous dual-fluorescent tracing mouse model [tyrosine kinase (Tek)–Cre;mTmG mice] with a modified 3D tissue-transparency technique, followed by scanning the transparent tissues under a high-resolution spinning-disc confocal microscope.
Tissues collection
- Perfusion with cold PBS to remove the blood (perform this step selectively depending on the content of blood in the tissues).
- Collect the tissues in 4% PFA in the dark for 24 hours at 4℃.
Clearing method of tissues (modified according to Li et al., Proc. Natl. Acad. Sci. USA. 2017 Aug 29;114(35):E7321-E7330. doi: 10.1073/pnas.1708981114.)
- Stock N-methylacetamide (M26305, Sigma-Aldrich) was prepared by diluting melted N-methylacetamide to 40% (v/v) in phosphate-buffered saline (PBS), which was then used to dissolve Histodenz (D2158, Sigma-Aldrich) to 86% (w/v) concentration.
- Triton X-100 (0.1% v/v) and 1-thioglycerol (M1753, Sigma-Aldrich) (0.5% v/v) were added to the Histodenz solution to be the final clearing solution.
- The tissues were collected and washed with PBS containing 0.2% Triton X-100 and 1-thioglycerol (0.5%) in the dark for 24 hours at room temperature to remove blood cells.
- The tissues were placed in the clearing medium (1:50 v/v) and incubated in the dark at room temperature on a rotor for 72 hours.
High-resolution 3D tissue imaging
- The cleared tissues were embedded in a 35-mm dish with 14-mm glass bottom (D35-14-1-N, Cellvis) containing fresh clearing solution and tightly covered by a coverslip.
- Confocal imaging was performed on an inverted Leica (DMi8) and Andor Dragonfly spinning-disc confocal microscope, a scientific complementary metal-oxide semi-conductor (sCMOS) camera (Andor Zyla 4.2), and using either a 20× 0.8 numerical aperture (NA) 650-μm working distance or a 40× 1.3 NA 250-μm working distance objective.
- A pixel density of 2048 × 2048 was used, and Z-step 0.5 to 1.0 μm for 650 μm (20× objective) or 0.3 to 0.6 μm for 250 μm (40× objective). The 488-nm (mG) and 568-nm mTomato (mT) lines of the Andor Integrated Laser Engine (ILE) system with a spinning-disc confocal scan head (Andor Dragonfly 500) were imaged. Images were acquired by Fusion 2.1 software (https://andor.oxinst.com/products/dragonfly#fusion).
Image processing
- Images were processed by ImageJ (http://rsbweb.nih.gov/ij/) for projection of all z stacks and merged color channels.
- The 3D vascular structure of tissues and the rotary 3D movie was processed by Imaris (https://imaris.oxinst.com/) software.
Materials
- 35-mm dish with 14-mm glass bottom (D35-14-1-N, Cellvis)
- N-methylacetamide (M26305, Sigma-Aldrich)
- Histodenz (D2158, Sigma-Aldrich)
- 1-thioglycerol (M1753, Sigma-Aldrich)
Images
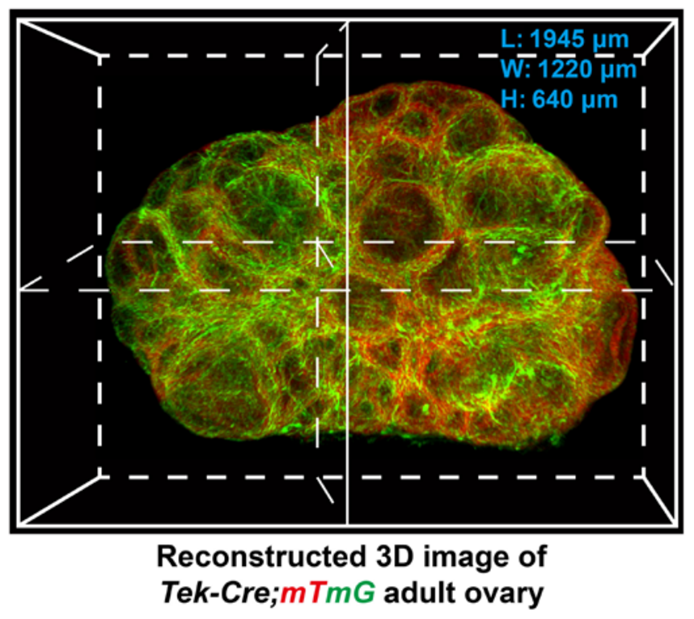
Xu et al., Sci Adv. 2022 Jan 14;8(2):eabi8683. doi: 10.1126/sciadv.abi8683.
The transparent ovaries were scanned by high-resolution confocal microscopy to reconstruct a 3D image of the ovarian blood vessels.
- Zhang, H, Mu, L, Xu, X, Yang, X and Zhang, Y(2023). High-resolution 3D tissue imaging. Bio-protocol Preprint. bio-protocol.org/prep2368.
- Xu, X., Mu, L., Li, L., Liang, J., Zhang, S., Jia, L., Yang, X., Dai, Y., Zhang, J., Wang, Y., Niu, S., Xia, G., Yang, Y., Zhang, Y., Cao, Y. and Zhang, H.(2022). Imaging and tracing the pattern of adult ovarian angiogenesis implies a strategy against female reproductive aging. Science Advances 8(2). DOI: 10.1126/sciadv.abi8683
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
