Advanced Search
Genome editing by CRISPR-Cas9
Last updated date: Mar 2, 2020 Views: 1246 Forks: 0
Endogenous tagging of LC3B with tandem GFP-mCherry in H4 cells using CRISPR-Cas9
Generation of LC3B-gRNA plasmid.
1. Synthesize oligonucleotides encoding LC3B-targeting gRNAs (targeting sequences highlighted in red).
Forward: 5'-CACCGTTCTCCGACGGCATGGTGCA-3’
Reverse: 5'-CTGCACCATGCCGTCGGAGAACAAA-3’
2. Mix 10 μl forward oligonucleotide and 10 μl reverse oligonucleotide (100 μM each in TE). Anneal in a thermocycler by heating at 95°C for 5 min and ramping down to 4°C at 0.1°C/sec. Dilute the annealed oligos at 1:200 with double-distilled H2O.
3. Digest 0.5 μg of pSpCas9 (BB)−2A-GFP plasmid (Addgene, 48138) with a mix of 1 μl BbsI (NEB, R0539), 2 μl NEBuffer™ 2.1 and double-distilled H2O to 20 μl for 4 h at 37°C.
4. Purify digested plasmid using NucleoSpin® Gel and PCR Clean-up kit (MACHEREY-NAGEL).
5. Ligate oligonucleotides into digested plasmid by mixing 50 ng digested plasmid, 3 μl diluted oligonucleotides, 5 μl DNA Ligation Kit, Mighty Mix (Takara, 6023), and double distilled H2O to 10 μl. Incubate at 25°C for 30 min.
6. Add 10 μl ligation mixture to 100 μl of NEB 5-alpha Competent E. coli (NEB, C2987). Mix gently by flicking the tube 4 or 5 times. Place the mixture on ice for 30 min. Heat shock at 42°C for 30 sec. Add 1,000 μl SOC medium (ThermoFisher Scientific, 15544034) to the bacteria and grow in 37°C shaking incubator for 1 h. Spread 100 μl of the bacteria onto a 10-cm LB agar plate containing the appropriate antibiotic. Incubate overnight at 37°C.
Generation of donor DNA plasmid for homologous recombination
7. Synthesize the following oligonucleotide primers:
| Primer | Sequence | PCR template | Product length |
| Left arm-forward | cactataggctagcctcgagCCAAATATCGCATGGTGGTTTACGCACTGC | H4 genomic DNA | 1,030 bp |
| Left arm-reverse | ccagctctgcGGTGCAGGGATCTGGGCGGC | ||
| GFP-mCherry-forward | tccctgcaccGCAGAGCTGGTTTAGTGAACCGTCAG | GFP-mCherry-LC3B plasmid | 1,661 bp |
| GFP-mCherry-reverse | cgacactcacCGAAGGTGCGGCGCTGCTTG | ||
| Right arm-forward | cgcaccttcgGTGAGTGTCGCCGCGAGGGC | H4 genomic DNA | 1,030 bp |
| Right arm-reverse | aagggaagcggccgcccgggTAGACTCCGGGAAACTAGGCAAAAGCAGAC |
8. PCR-amplify each fragment according to the following conditions by using Q5® High-Fidelity 2X Master Mix (NEB, M0492):
| Step | Temperature | Time |
| Initial denaturation | 98°C | 30 sec |
| 35 cycles | 98°C 50–72°C* 72°C | 10 sec 20 sec 20 sec/kb |
| Final extension | 72°C | 2 min |
| Hold | 4°C |
*Annealing temperature is calculated by using NEB Tm Calculator (https://tmcalculator.neb.com/).
9. Digest 0.5 μg of pCI-neo plasmid (Promega) with EcoRI and SalI for 4 h at 37°C.
10. Gel-purify PCR product and digested plasmid using NucleoSpin® Gel and PCR Clean-up kit (MACHEREY-NAGEL).
11. Perform Gibson assembly by mixing 50 ng digested pCI-neo, 50 ng left arm, 50 ng right arm, 85 ng GFP-mCherry (the molarity ratio of pCI-neo and other three fragments are 1:5:5:5) in 10 μl Gibson Assembly® Master Mix (NEB, E5510) and double-distilled H2O to 20 μl. Incubate at 50°C for 30 min.
12. Transform E. coli as described above.
Generation of the H4 tandem fluorescent LC3B (H4-tfLC3B) cell line
13. H4 cells were grown in Dulbecco's modified Eagle's medium (DMEM, Corning, 15-013-CV) with 10 % fetal bovine serum (FBS, Corning, 35-011-CV), 100 IU/ml penicillin, 100 μg/ml streptomycin (Corning, 30-002-CI) and 2 mM L-glutamine (Corning, 25-005-CI) at 37°C, 5 % CO2.
14. Seed 1.5 x 106 H4 cells in a 10-cm tissue culture dish one day before transfection.
15. Dilute 16 μl Lipofectamine® 2000 reagent (Thermofisher Scientific, 11668019) in 250 μl Opti-MEM® medium (Thermofisher Scientific, 31985070).
16. Dilute 2 μg LC3B-gRNA plasmid and 2 μg donor DNA plasmid in 250 μl Opti-MEM® medium.
17. Add diluted DNA to diluted Lipofectamine® 2000 reagent. Incubate for 5 min at room temperature. Add DNA-lipid complex to cells.
18. One hour after transfection, aspirate transfection medium and replace with fresh medium.
19. After 24 h, sort GFP-positive cells on a FACS Aria II Flow Cytometer (BD Biosciences), and propagate cells in regular culture medium.
20. One week after transfection, sort GFP-mCherry double-positive cells by FACS, and propagate cells in regular culture medium.
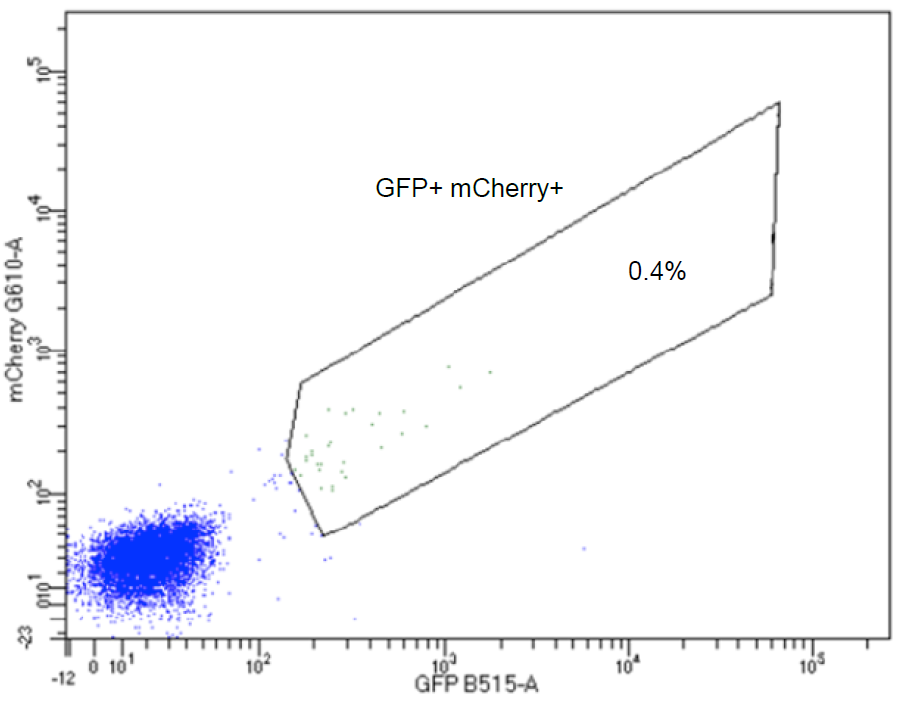
Dot plot indicating the GFP-mCherry double-positive H4 cells in the boxed area. Gated cells are collected for propagation and monoclonal cell isolation.
21. Isolate monoclonal cell populations by limiting dilution in a 96-well plate. Trypsinize the stable cell pool and break up any cell clumps by passing several times through a serological pipet. Quantify the cell concentration. Dilute the cell suspension with culture medium to a concentration of 5 cells/ml. Transfer 100 μl of the suspension into each well of a 96-well plate.
22. After the monoclonal lines have been sufficiently expanded (it normally takes 2 weeks), trypsinize the cells in the 96-well plate and divide the cells from each well in two parts: one part to be seeded in a well of a 24-well plate for further expansion, the other part for genomic DNA extraction using QuickExtract™ DNA Extraction Solution (Epicentre,QE09050).
23. Validate the clonal cell lines by PCR using the following conditions:
Primers:
| Primer | Sequence | Targeting site | Product length |
| LC3B-validate-forward | CGTGAACGGCCACGAGT | mCherry | 2,734 bp |
| LC3B-validate-reverse | CACTCAAGCTAACTGGGGCTT | First intron of LC3B gene |
PCR reaction:
| Step | Temperature | Time |
| Initial denaturation | 98°C | 60 sec |
| 40 cycles | 98°C 68°C 72°C | 10 sec 20 sec 60 sec |
| Final extension | 72°C | 2 min |
| Hold | 4°C |
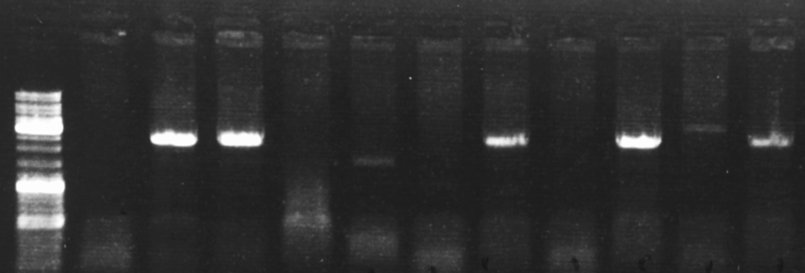
Representative DNA electrophoresis gel showing 5 out of 11 single-cell clones that are successfully edited.
24. Validate theclonal cell lines by immunoblotting.
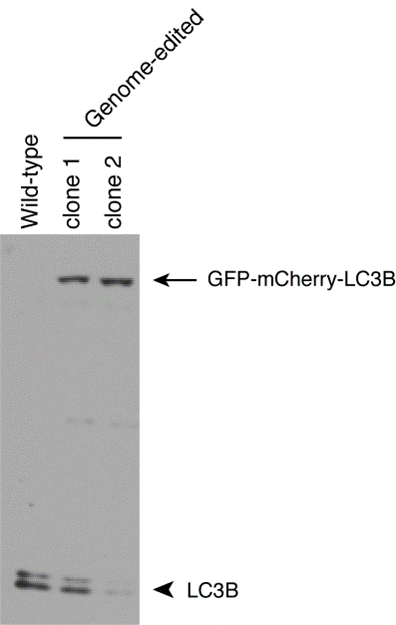
Wild-type H4 cells and two genome-edited single-cell clones are analyzed by immunoblotting with antibody to LC3B (Cell Signaling Technology, 3868). The arrow indicates GFP-mCherry-tagged LC3B; the arrowhead indicates untagged LC3B.
25. Validate the clonal cell lines by FACS.
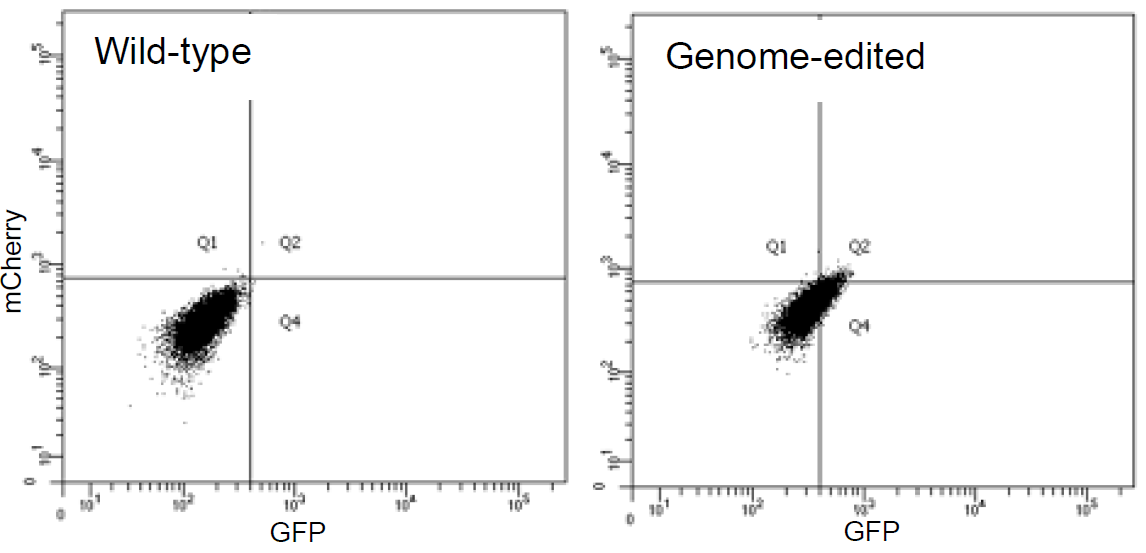
Wild-type and genome-edited H4 cells are analyzed by FACS. The population of genome-edited H4 cells shifts to the GFP-mCherry double-positive quadrant (Q2), indicating that both GFP and mCherry are expressed in this cell line.
26. Validate the clonal cell lines by confocal fluorescence microscopy.
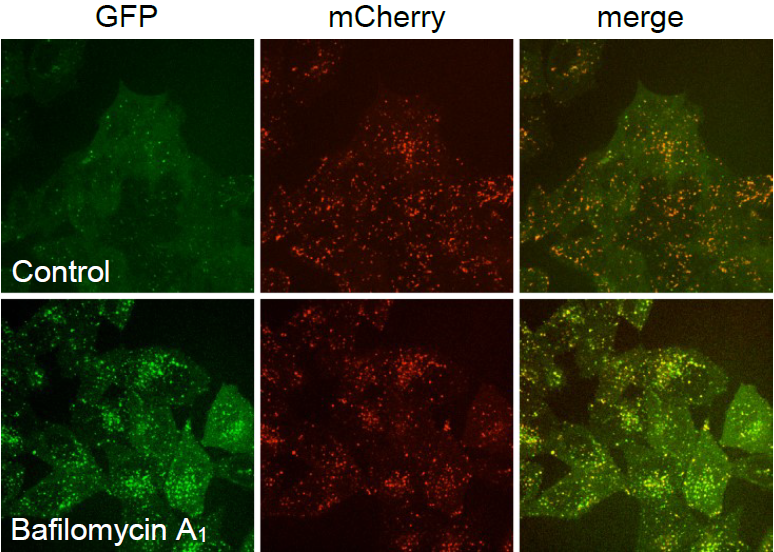
Live-cell imaging of genome-edited H4 cells incubated with vehicle (Control) or 50 nM bafilomycin A1 (Sigma-Aldrich, B1793) for 2 h. At the neutral pH of autophagosomes, both the GFP and mCherry fluoresce. After the fusion of autophagosomes with lysosomes, GFP is quenched in the acidic autolysosomes, and only the mCherry fluorescence is visible. Bafilomycin A1 inhibits acidification and lysosomal degradation, and GFP fluorescence is no longer quenched in autolysosomes, leading to the increase of GFP signal in mCherry-positive autolysosomes.
Generation of UBA6-KO and BIRC6-KO H4 cells using CRISPR-Cas9
1. Synthesize oligonucleotides encoding gRNAs targeting UBA6 or BIRC6. Two gRNAs are designed for each gene (targeting sequences are highlighted in red).
UBA6-1-forward: 5'-CACCGCGAGCCTGTGGCCGCCCATC-3'
UBA6-1-reverse: 5'-CGATGGGCGGCCACAGGCTCGCAAA-3'
UBA6-2-forward: 5'-CACCGCTCCGGTCGAGAGCGAGTTC-3'
UBA6-2-reverse: 5'-CGAACTCGCTCTCGACCGGAGCAAA-3'
BIRC6-1-forward: 5'-CACCGGCATGCACTGCGACGCCGAC-3'
BIRC6-1-reverse: 5'-CGTCGGCGTCGCAGTGCATGCCAAA-3'
BIRC6-2-forward: 5'-CACCGTCTCGCTTCCCCGAGTCGCG-3'
BIRC6-2-reverse: 5'-CCGCGACTCGGGGAAGCGAGACAAA-3'
2. Anneal each pair of oligonucleotides and clone them into pSpCas9 (BB)−2A-GFP plasmid as described above.
3. Co-transfect H4 cells with the two plasmids containing the different targeting sequences for the same gene as described above.
4. Sort GFP-positive cells by FACS and isolate monoclonal cell populations by limiting dilution in a 96-well plate as described above.
5. Validate the cleavage of the target sequence in the monoclonal cell lines by PCR.
| Primer | Sequences | Product length |
| UBA6-validate-forward | CGTGAACGGCCACGAGT | WT: 772 bp KO: ~420 bp |
| UBA6-validate-reverse | CACTCAAGCTAACTGGGGCTT | |
| BIRC6-validate-forward | ACTTCACTTCCGGCTAACGC | WT: 738 bp KO: ~500 bp |
| BIRC6-validate-reverse | CTCATCTAACCGGACGCAGG |
- Jia, R and Bonifacino, J(2020). Genome editing by CRISPR-Cas9. Bio-protocol Preprint. bio-protocol.org/prep233.
- Jia, R. and Bonifacino, J. S.(2019). Negative regulation of autophagy by UBA6-BIRC6–mediated ubiquitination of LC3. eLife. DOI: 10.7554/eLife.50034
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
