Advanced Search
Numerical methods
Last updated date: May 26, 2023 Views: 960 Forks: 0
We modeled the long-term dynamics and diversity of ecological communities using the well-known generalized Lotka-Volterra (gLV) model, modified to include dispersal from a species pool:
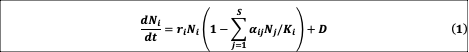
where Ni is the abundance of species i (normalized to its carrying capacity), αij is the interaction strength that captures how strongly species j inhibits the growth of species i (with self-regulation αii = 1), and D is the dispersal rate from an outside species pool to the focal community. For simplicity and without qualitatively changing our results, we considered the same growth rate
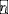
= 1 and the same carrying capacity
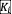
= 1 for all species in the main text. Fig. S6 shows that sampling growth rates from a uniform distribution has little effect on the phase diagram of survival fraction and fluctuation fraction. Fig. S6 shows that sampling carrying capacities from a normal distribution increases the partial coexistence phase while shrinking both the full coexistence phase and fluctuation phase, but does not affect the order of the phases. We further tested the theoretical predictions when considering the existence of positive (facilitative) interspecies interactions (Fig. S9) and varying the symmetry of the interaction matrix (Fig. S8). We also considered different dispersal rates (Fig. S7), and the effects of incorporating daily dilutions (Fig. S9) in these in silico communities. These additional results show that our qualitative phase diagrams and conclusions are robust to different choices of ecological network structure and parameters. Although the patterns of ecological diversity and dynamics do not change as the dispersal rate varies from D=10-7 to D=10-6 (Fig. S7), we found that communities with zero dispersal rate exhibit lower fluctuation fraction and survival faction in the persistent fluctuation phase. Our results show that non-zero dispersal rates can sustain persistent fluctuations.
To show that symmetry and anti-symmetry do not qualitatively change the phase diagram, we simulated communities in two scenarios of non-zero reciprocity,
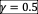
and

, where the reciprocity of interactions is given by
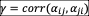
. The qualitative patterns and order of transitions in the phase diagram (Fig. S8) are robust to the presence of reciprocity, although
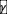
shifts the stability boundary (solid line in Fig. S8). Positive and negative reciprocity decreases and increases the values of S and <
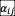
> at which communities lose stability, respectively. Moreover, positive (negative) reciprocity yields lower (higher) survival fraction and fluctuation fraction of communities. At full symmetry and anti-symmetry (
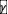
=1, -1) there is no fluctuating phase (21), but those do not appear to be relevant from the experiment pair-competition results (Table. S1-S3).
To test whether the existence of positive (facilitative) interactions in the ecological network will change our conclusions, we sampled values of
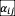
from a uniform distribution [-
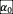
,
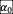
], where
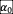
varies between [0, 1.4] on the phase diagram. In this simulation, the linear interaction function in the gLV (

) is replaced with Monod function (

) to avoid unbounded growth due to positive interactions (21, 42). We observed similar patterns of survival fraction and fluctuation fraction between pure competitive interactions and considering positive interactions (Fig. S9). The first and second moment of the distribution of
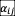
should be considered to quantify interaction strength in the stability criteria (17). Here we use Std (
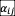
) to quantify the interaction strength because the first moment of
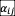
distribution is zero. Our results demonstrate that the existence of three phases (full coexistence, partial coexistence, persistence fluctuation) and the order of transitions are robust to the interaction types in the model.
All simulations used the Runge-Kutta method on Matlab to numerically solve the LV equations (with an integration step of 0.05). A definition of 100
100 pixels was used for each phase diagram, linearly segmenting the parameter space in the ranges <αij> ∈ [0, 1.5] and S ∈ [1, 100]. In each phase diagram, each pixel shows the average result for 103 simulations. The total simulation time is 104 to guarantee the survival fraction and fluctuation fraction have reached steady states as shown in Fig. S5.
To test whether the total biomass fluctuation is consistent with species abundance fluctuation in our simulations, we simulated the time series of community biomass under various conditions (Fig. S3). To quantify the dynamics of biomass in the simulation, we calculated the sum of species abundance (

). Our results demonstrate the fluctuation in species abundance is in agreement with fluctuation in total biomass.
The similar fluctuations between replicates in some experiments (e.g., Fig. 2, medium nutrients, S=48) could be explained by the slow divergence that chaotic trajectories can exhibit during moderately long-time windows, or, alternatively, by possible limit cycle oscillations (Fig. S3). We focused on chaotic fluctuations when discussing the model predictions because previous theory shows that all persistent fluctuations will be chaotic as number of species in the pool S grows (21), though limit cycle oscillations only happen under finite S.
Below are the Matlab code:
clear;clc;close all;
r_mean=1.0; % mean growth rate
S=1:1:100; % number of species
DD=1e-6; % migration rate
A_mean_range=[0.01 1.5]; % mean inhibition alpha
ystep=100; %for A_mean
T_real=10000; %total evolution time
step=0.05; % time interval
T=ceil(T_real/step); %number of circulation
num_sim=10000; % number of simulation
period=2000; % duration for calculating richness
% composition_full=zeros(length(S)*ystep*num_sim,length(S)); % record the final compositions of all simulations
abundance_threshold=1e-3; %threshold for survival species
fluc_record=zeros(length(S)*ystep,num_sim);
diversity_record=zeros(length(S)*ystep,num_sim);
A_mean=linspace(A_mean_range(1),A_mean_range(2),ystep);
for mm=1:length(S)
for pp=1:ystep
for hh=1:num_sim
composition= LV_compute(r_mean, S(mm), DD, A_mean(pp),T,step);
diversity_index=max(composition(T-period+1:T,:)); richness=find(diversity_index>abundance_threshold);
diversity_record((mm-1)*ystep+pp,hh)=length(richness)/S(mm);
fluc_CV=std(composition(T-1e5:T,:))./mean(composition(T-1e5:T,:)); fluc_CV(isnan(fluc_CV))=0;
fluc_record((mm-1)*ystep+pp,hh)=mean(fluc_CV(fluc_CV>0));
end
end
end
function [composition] = LV_compute(r_mean, S, D, A, T, step)
N_mean=1/(S*A*sqrt(pi/2));
sigma3=N_mean/12;
NN=zeros(T,S);
r=ones(S,1);
% Interaction matrix
% AA=exprnd(A,S,S);
AA=rand(S,S)*A*2;
AA=AA-diag(diag(AA))+eye(S);
% considering carrying capacity
% K=normrnd(1,sqrt(1/12),[S,1]);
% for i=1:S
% for j=1:S
% AA(i,j)=AA(i,j)*K(j)/K(i);
% end
% end
% Initial composition
N0=abs(normrnd(N_mean,sigma3,1,S));
NN(1,:)=N0;
% Migration from source
Migra=ones(1,S)*D;
% compute the dynamics in each patch
for i=2:T
for j=1:S
k1=r(j)*NN(i-1,j)*(1-AA(j,:)*(NN(i-1,:)'))*step;
k2=r(j)*(NN(i-1,j)+k1/2)*(1-AA(j,:)*(NN(i-1,:)'))*step;
k3=r(j)*(NN(i-1,j)+k2/2)*(1-AA(j,:)*(NN(i-1,:)'))*step;
k4=r(j)*(NN(i-1,j)+k3)*(1-AA(j,:)*(NN(i-1,:)'))*step;
NN(i,j)=NN(i-1,j)+(1/6)*(k1+2*k2+2*k3+k4);
end
NN(i,:)=NN(i,:)+Migra;
end
composition=NN;
end
clear; clc; close all;
load ./uniform3.mat;
load feasible_exp.mat
x_step=length(S); y_step=length(A_mean);
miu1=A_mean(1); miu2=A_mean(length(A_mean)); S1=S(1); S2=S(length(S));
color12=[0.7 0.7 0.7];
phase_indicate=mean(diversity_record,2); fluc_digi=fluc_record;
phasenew_indicate=zeros(length(phase_indicate),1);
critical_CV=0.001;
for i=1:length(phasenew_indicate)
for j=1:num_sim
if fluc_record(i,j)<critical_CV
fluc_digi(i,j)=0;
else
fluc_digi(i,j)=1;
end
end
end
fluc_indicate=mean(fluc_digi,2);
phase_std=std(diversity_record,0,2)./phase_indicate;
phasecolor=reshape(phase_indicate,[y_step,x_step]); phasecolor_std=reshape(phase_std,[y_step,x_step]);
phasecolor_fluc=reshape(fluc_indicate,[y_step,x_step]); phasecolor_new=reshape(phasenew_indicate,[y_step,x_step]);
phasecolor_new1=zeros(y_step,x_step);
x2=linspace(miu1,miu2,y_step)'; x1=sqrt(linspace(S1,S2,x_step))'; x2(1)=0;
X1=repmat(x1',y_step,1); X2=repmat(x2,1,x_step);
figure(1)
s=pcolor(X1.^2,X2,phasecolor);set(s, 'LineStyle','none'); colorbar; hold on;
caxis([0.0 1.1])
plot(xx3,yy3,'k-','linewidth',5,'color',color12)
plot(ss,fea,'k-.','linewidth',5,'color',color12);
axis([S1 S2 0 1.5]); set(gca,'FontSize',28);
xlabel('Size of species pool, {\itS}','FontSize',30); ylabel('Interaction strength, <\it\alpha_i_j>','FontSize',30);
xticks([1 20 40 60 80 100]); yticks([0.0 0.5 1.0 1.5]); box on
figure(2)
p=0.9;
sp=csaps({x1.^2,x2},phasecolor_fluc,p);
v=fnval(sp,{x1.^2,x2});
s=pcolor(X1.^2,X2,v);set(s, 'LineStyle','none'); colorbar; hold on;
caxis([0.0 1])
plot(xx3,yy3,'k-','linewidth',5,'color',color12)
plot(ss,fea,'k-.','linewidth',5,'color',color12);
axis([S1 S2 0 1.5]); set(gca,'FontSize',28);
xlabel('Size of species pool, {\itS}','FontSize',30); ylabel('Interaction strength, <\it\alpha_i_j>','FontSize',30);
xticks([1 20 40 60 80 100]); yticks([0.0 0.5 1.0 1.5]); box on
- Hu, J and Gore, J(2023). Numerical methods. Bio-protocol Preprint. bio-protocol.org/prep2323.
- Hu, J., Amor, D. R., Barbier, M., Bunin, G. and Gore, J.(2022). Emergent phases of ecological diversity and dynamics mapped in microcosms. Science 378(6615). DOI: 10.1126/science.abm7841
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
