Advanced Search
Expression and purification of CR3022 Fab and IgG
Last updated date: Apr 4, 2023 Views: 106 Forks: 0
Expression and purification of SARS-CoV-2 receptor binding domain and CR3022 antibody
Meng Yuan1,*, Nicholas C. Wu1,$, Wenli Yu1, and Ian A. Wilson1,2
1 Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA,
2 The Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
$ Present address: Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
* For correspondence: myuan@scripps.edu
Abstract
The SARS-CoV-2 receptor binding domain (RBD) is a major target for neutralizing antibodies. Here we report our method for production of the SARS-CoV-2 RBD that has been extensively used in numerous studies. Although the RBD of SARS-CoV-2 is variable among variants and other sarbecoviruses, some regions on the RBD are highly conserved, including the CR3022 site. We also present a detailed protocol for production of the IgG and Fab of antibody CR3022, which cross-reacts with a broad range of sarbecoviruses, including SARS-CoV-1 and SARS-CoV-2. The production of SARS-CoV-2 RBD and CR3022 antibody provides valuable materials for COVID-19 research.
Key Features
- Molecular cloning, protein expression and purification of the SARS-CoV-2 receptor binding domain
- Molecular cloning, protein expression and purification of CR3022 IgG
- Molecular cloning, protein expression and purification of CR3022 Fab
Keywords: SARS-CoV-2, COVID-19, coronavirus, antibody, CR3022
Background
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants have resulted in enormous global health and socioeconomic damage and requires the development of effective vaccines and broadly neutralizing therapeutic antibodies. Early in the pandemic, we and others found that an antibody previously isolated from a patient infected with SARS-CoV-1, CR3022, cross-reacted with the SARS-CoV-2 receptor binding domain (RBD) (Huo et al., 2020; ter Meulen et al., 2006; Yuan et al., 2020). We determined a crystal structure of the SARS-CoV-2 RBD in complex with CR3022, revealing a cryptic site that is highly conserved among sarbecoviruses (Yuan et al., 2020). Besides the CR3022 site, we also initially defined the SARS-CoV-2 RBD into six major epitope regions, including four RBS sites (RBS-A, -B, -C, and -D), S309 site, as well as the CR3022 site (Yuan et al., 2021a; Yuan et al., 2021b). Another site V has also recently emerged (Starr et al., 2021). Although the RBD is highly variable, especially in the RBS sites, the CR3022 site remains relatively conserved among SARS-CoV-2 variants and other sarbecoviruses (Yuan et al., 2022). Here, we present protocols for robust production of wild-type SARS-CoV-2 RBD and CR3022 antibody (IgG and Fab). The method is also transferable to RBDs of other variants. Similarly, the production of CR3022 antibody can be applied to other human antibodies. The protocols for production of SARS-CoV-2 RBD and antibodies have aided research on COVID-19.
Materials and Reagents
Biological Materials
- Sf9 cells
- High Five cells
- Expi293TM cells (ThermoFisher Scientific, catalog number: A14527)
- DH5α competent cells
- DH10Bac competent cells (Invitrogen, 10359-016)
Reagents
- pFastBac vector pre-constructed with gp67 signal peptide (5'-ATGGTACTAGTAAATCAGTCACACCA AGGCTTCAATAAGGAACACACAAGCAAGATGGTAAGCGCTATTGTTTTATATGTGCTTTTGGCGGCGGCGGCGCATTCTGCCTTTGCGGCGGATCCCGGG-3')
- Primers for linearizing the pFastBac vector for PIPE cloning of the SARS-CoV-2 receptor binding domain (RBD) (5'-CCCGGGATCCGCCGCAAAGGCAGAATGC-3' and 5'-AGCGGCCATCACCACCATCACC ATTGAGGCCAGGCCGGCCGCATGCG-3')
- Human codon optimized synthetic RBD gene (encoding amino acids 319-541 of SARS-CoV-2 Spike protein, GenBank: QHD43416.1) with 15-bp extensions overlapping with pFastBac in-fusion entry sites.
- phCMV3 vector pre-constructed with human antibody constant domains and signal peptide (5'-ATGGAGACAGACACACTGCTGCTGTGGGTGCTGCTGCTGTGGGTGCCAGGCAG
CACCGGC-3'). - Primers for linearizing the phCMV3 vector for PIPE cloning of CR3022 IgG and Fab
| Forward | Reverse | |
IgG heavy chain | 5'-CAAGGGACCAAGCGTGTTCCCACTG-3' | 5’-TGCCTGGCACCCACAGCAGCAGCAC-3' |
Fab heavy chain | 5'-CAAGGGACCAAGCGTGTTCCCACTG-3' | 5’-TGCCTGGCACCCACAGCAGCAGCAC-3' |
Kappa light chain | 5'-GGCAGCACCTTCCGTGTTCATCTTT-3' | 5’-TGCCTGGCACCCACAGCAGCAGCAC-3' |
6. Human codon optimized synthetic CR3022 genes (GenBank: DQ168569.1 and DQ168570.1 for heavy and light chains, respectively) with 15-bp extensions overlapping with phCMV3 in-fusion entry sites.
7. KOD polymerase (MilliporeSigma, 710863)
8. DpnI (NEB, R0176L)
9. Exonuclease III (Takara, 2170A)
10. S.O.C. medium (ThermoFisher, 15544034)
11. FuGENE® HD Transfection Reagent (Promega, E2311)
12. Insect-XPRESSTM Protein-free Insect Cell Medium (Lonza, 12-730Q)
13. Ni Sepharose excel histidine-tagged protein purification resin (Cytiva, 17371203)
14. Superdex200 pg (GE: 17-1043-02)
15. Imidazole (MillieporeSigma, 12399-500G)
16. Dulbecco's Phosphate-Buffered Saline (Corning, 21-031-CM)
Laboratories Common Solution
- 1% agarose gel in 1× TAE buffer and 1× TAE running buffer
- 1× SDS protein Gel running buffer
- 0.5 M EDTA (pH 8.0)
- LB medium
- Tris-buffered saline (TBS), pH=7.4
- Phosphate-buffered saline (PBS), pH=7.4
- 50 mM acetate buffer, pH 4.0
Laboratory Supplies
- QIAprep Spin Miniprep Kit (QIAprep, 27104)
- PureLink HiPure Plasmid Miniprep Kit (ThermoFisher, K210002)
- NucleoBond Xtra Midi Plus Kit (Takara, 740412.50)
- T25 cm2 flask (Corning, 430639)
- T-175 flask (Corning, 431080)
- 3 L Flask (Corning, 431252)
- Centramate cassette (PALL Life Sciences, OS010T12)
- Econo column (Bio-Rad, 7372512)
- Amicon® centrifugal filters (Amicon, UFC801096)
- Hiload 16/90 Superdex® 200 column (GE Healthcare)
- Superdex® 200 Increase 10/300GL (Cytiva, 28990944)
- Mini-protein TGX Stain-Free Gel (Bio-Rad, 4568093)
Equipment
- PCR (Bio-rad, C1000 Touch Thermal Cycler)
- Lab pump (Millipore, XX8200115)
- HPLC (Cytiva, AKTA pure)
- Centrifuges (Beckman, Allegra X-22R; Beckman, Microfuge 18)
- Nanodrop (ThermoScientific, ND-1000 Spectrophotometer)
- CO2 orbital shaker (Infors HT, Multitron Pro)
- Mini-PROTEAN® Tetra Cell and PowerPac™ HC Power Supply (Bio-Rad, 1658027)
- Mini-protean TGX Gel (Bio-Rad, 4568093)
Procedure
A. Production of SARS-CoV-2 receptor binding domain
- Plasmid construction
a. Perform PCR to linearize pFastBac vector:
5 µl 10× KOD Hot-start buffer
3 µl 25 mM MgSO4
5 µl 2 mM dNTP mix
1.5 µl primer 1 (10 µM)
1.5 µl primer 2 (10 µM)
1 µl KOD polymerase
5 µl 5 ng/µl DNA template
1 µl 100% DMSO
Add ddH2O to 50 µl
Run PCR:
Step 1: 95°C 2:00
Step 2: 95°C 0:20
Step 3: 63°C 0:15
Step 4: 68°C 30 sec/kbp
Go to step 2, run for 25 cycles
Step 5: 68°C 3:00
Step 6: 12°C storage
b. Add 1.5 µl of DpnI to the PCR product and incubate at 37°C for 2 h.
c. Run 1% agarose gel to visualize the DNA. Extract linearized DNA using a gel extraction kit by following the kit manual.
d. Measure the concentration of the produced DNA using a Nanodrop® Spectrophotometer.
e. PIPE cloning
Set Reaction system:
Vector 50-100 ng
Insertion 50-100 ng
10× ExoIII buffer 1 µl
Add ddH2O 10 µl
Place the reaction on an ice bath for 5 min. Add 1 µl of ExoIII and place it on an ice bath for 30 min. Neutralize the reaction by adding 1 µl of 0.5 M EDTA (pH=8.0). Inactivate the enzyme by incubating the reaction at 65°C for 5 min. Anneal the reaction by incubating at 4°C for 5 min.
f. Transformation
Add 1 µl of the PIPE cloning solution to 25 µl of DH5 α competent cells. Incubate the mixture on ice for 10 min. Heat shock the cells at 42°C for 45 seconds. Immediately place the tube on ice and incubate for 2 min. Add 250 µl of SOC medium to the tube and shake it at 37°C, 200–300 rpm, for 2 h. Streak the transformed bacteria on an LB plate containing gentamycin. Incubate the plate overnight at 37°C. Use another plate containing only linearized vector transformed bacteria as a control experiment.
g. Miniprep and DNA sequencing
Inoculate bacteria colonies into 3-mL tubes containing LB medium with gentamycin. Incubate the tubes overnight at 37°C with shaking at 200–300 rpm. Extract the plasmid DNA from the cell culture using a QIAprep Spin Miniprep Kit by following the manufacturer’s instructions. Sequence the extracted plasmid using primers: pFastBacF (5'-GGATTATTCATACCGTCCCA-3') and pFastBacR (5'-CAAATGTGGTATGGCTGATT-3'). - Construction of bacmid
- Add 0.5 μL of plasmid DNA to 50 μL of DH10Bac competent cells.
- Incubate the mixture on ice for 30 minutes.
- Heat shock the cells at 42°C for 30 seconds.
- Incubate on ice for 2 minutes.
- Add 300 μL SOC medium to the tube, shake at 220 rpm, 37ºC incubate for 4 h.
- Plate 100 μL of the mixture on KGXI plates (containing 50 μg/mL kanamycin + 15 μg/mL gentamycin + X-gal + IPTG), perform 1 to 10, 100, 1000 dilution.
- Wrapped in foil, incubate at 37°C for 48 hours.
- Pick a white colony and streak on a new plate. Incubate the plate at 37ºC for. Make sure all colonies are white.
- Extract the plasmid DNA from the cell culture using a PureLink HiPure Plasmid Miniprep Kit by following the manufacturer’s instructions.
- Transfection and baculovirus amplification
a. Seed 2×106 Sf9 cells into a T25 cm2 flask for each transfection with a total volume of 5 mL.
b. Make transfection mixture:
2 μg DNA bacmid
8 μL FuGene
1 mL Insect-XPRESS media
Mix well and incubate at room temperature for 20 min.
c. Add the transfection mixture to the surface of the Sf9 cells and shake the flasks to mix. Incubate at 27ºC for five days.
d. Harvest the virus from the P0 transfection.
e. Seed 15×106 Sf9 cells in a T-175 flask with a total volume of 50 mL. Infect the Sf9 cells with 0.25 mL of virus from the P0 transfection. Incubate at 27ºC with shaking at 110 rpm for five days. - Protein expression
- Seed 1×106 High Five cells/mL in a total volume of 1.5 liters, add 15 mL of P1 virus. Harvest virus after 72 hours.
- Allow to protein expression at 27ºC with shaking at 110 rpm for three days.
- Protein purification
- Centrifuge cells at 3,000× g for 0.5 h at 4ºC. Remove the cell pellet.
- Concentrate the supernatant using a 10 kDa MW cutoff Centramate cassette and buffer exchange into PBS.
- Add NiNTA resin to the supernatant, incubate the bottle on a bottle roller at 4ºC for 2 hours.
- Pour the liquid to an Econo column.
- Wash the resin using PBS, pH 7.4, followed by imidazole washing buffer (20 mM imidazole in PBS, pH=7.4).
- Elute with 250 mM imidazole in PBS, pH 7.4.
- Dialysis against TBS buffer (20 mM Tris, 150 mM NaCl, pH=7.4) overnight at 4°C.
- Concentrate elution to small volume using Amicon® centrifugal filters for loading onto an AKTA purification system.
- Run the sample through a size exclusion column (Superdex® 200) that is equilibrated with TBS buffer (20 mM Tris, 150 mM NaCl, pH=7.4) at 4ºC. Collect sample from the right peak (Figure 1)
- Concentrate the protein product to ~10 mg/mL using Amicon® centrifugal filters. Run SDS-PAGE to check protein quality (Figure 2A).
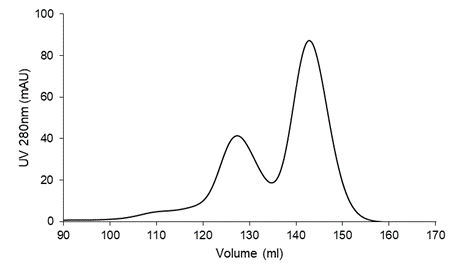
Figure 1. Purification of SARS-CoV-2 RBD on a Superdex® 200 16/90 size exclusion column.
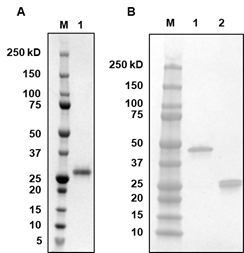
Figure 2. SDS-Page of (A) SARS-CoV-2 RBD and (B) CR3022 Fab. 5× reducing loading buffer that contains 0.5M of dithiothreitol (DTT) was used for A1 and B2, whereas 5× non-reducing loading buffer that contains no DTT was used for B1.
B. Production of CR3022 Fab and IgG
- Plasmid construction
a. Perform PCR to linearize phCMV3 vector:
5 µl 10× KOD Hot-start buffer
3 µl 25 mM MgSO4
5 µl 2 mM dNTP
1.5 µl primer 1 (5 µM)
1.5 µl primer 2 (5 µM)
1 µl KOD polymerase
5 µl 5 ng/µl DNA template
1 µl 100% DMSO
Add ddH2O to 50 µl
Run PCR:
Step 1: 95°C 2:00
Step 2: 95°C 0:20
Step 3: 63°C 0:15
Step 4: 68°C 30 sec/kbp
Go to step 2, run for 25 cycles
Step 5: 68°C 3:00
Step 6: 12°C storage
b. Add 1.5 µl of DpnI to the PCR product and incubate at 37°C for 2 h.
c. Run 1% agarose gel for the DNA and extract linearized DNA using a gel extraction kit by following the kit manual.
d. Measure the concentration of the produced DNA using a Nanodrop® Spectrophotometer.
e. PIPE cloning
Set Reaction system:
Vector 50-100 ng
Insertion 50-100 ng
10× ExoIII buffer 1 µl
Add ddH2O 10 µl
Place the reaction on an ice bath for 5 min. Add 1 µl ExoIII to the reaction in the ice bath for 30 min. Neutralize the reaction by adding 1 µl 0.5 M EDTA (pH=8.0). Inactivate the enzyme by heating the reaction at 65°C for 5 min. Anneal the reaction by incubating at 4°C for 5 min.
f. Transformation
Add 1 µl of the PIPE cloning solution, add to 25 µl of DH5 α competent cells, incubate on ice for 10 min. Heat shock 42°C for 45 seconds. Put on ice and incubate for 2 min. Add 250 µl of SOC medium, shake at 37°C at 200–300 rpm for 1 h. Streak on an LB plate with kanamycin resistance. Incubate the LB plate at 37°C overnight. Another plate with only linearized vector transformed bacteria is used as control experiment.
g. Miniprep and DNA sequencing
Inoculate bacteria colonies into 3-mL tubes containing LB medium with kanamycin. Shake at 200–300 rpm at 37°C overnight. Extract the plasmid from the cell culture using a DNA plasmid miniprep kit by following the manual instructions. Sequence the extracted plasmid using primers: T7 (5'- TAATACGACTCACTATAGGG-3') and pFastBacR (5'-CAAATGTGGTATGGCTGATT-3'). - DNA plasmid amplification
a. Transformation
Add 1 µl of the DNA plasmid, add to 25 µl of DH5 α competent cells, incubate on ice for 10 min. Heat shock 42°C for 45 seconds. Put on ice and incubate for 2 min. Add 250 µl of SOC medium, shake at 37°C at 200–300 rpm for 1 h. Streak on an LB plate with kanamycin resistance. Incubate the LB plate at 37°C overnight.
b. Bacteria culture
Pick a single colony in the LB plate and inoculate into 100 mL LB medium with kanamycin. Shake at 200–300 rpm at 37°C overnight. Extract the DNA plasmid from the cell culture using a NucleoBond Xtra Midi Plus Kit by following the manual. - Protein expression
- Transfect the CR3022 antibody DNA plasmids (66.7 µg and 33.3 µg for heavy and light chains, respectively) into 100 mL of Expi293FTM or ExpiCHOTM cells following the corresponding manuals.
- For expression with Expi293FTM cells, shake at 125 rpm at 37°C for 6 – 7 days. For expression with ExpiCHOTM cells, shake at 125 rpm at 32°C for 12 – 14 days.
- Protein purification
- Centrifuge cells at 3,000× g for 1 h at 4ºC. Remove the cell pellet.
- Add CaptureSelectTM CH1-XL resin to the supernatant, incubate the bottle on a bottle roller at 4ºC for 2 hours.
- Pour the liquid to an Econo column.
- Wash the resin using 100 mL PBS, pH 7.4.
- Elute with 15 mL of 50 mM acetate buffer, pH 4.0.
- Dialysis against TBS buffer (20 mM Tris, 150 mM NaCl, pH=7.4) overnight at 4°C.
- Concentrate elution to small volume using Amicon® centrifugal filters for loading onto an AKTA purification system.
- Run the sample through a size exclusion column (Superdex® 200 16/90) that is equilibrated with TBS buffer (20 mM Tris, 150 mM NaCl, pH=7.4) at 4ºC.
- Concentrate the protein product using Amicon® centrifugal filters. Run SDS-PAGE to check protein quality (Figure 2B).
Acknowledgements
This work was supported by the Bill and Melinda Gates Foundation OPP1170236 (I.A.W.)
Competing interests
N.C.W. consults for HeliXon. The authors declare no other competing interests.
References
Huo, J., Zhao, Y., Ren, J., Zhou, D., Duyvesteyn, H. M. E., Ginn, H. M., Carrique, L., Malinauskas, T., Ruza, R. R., Shah, P. N. M., Tan, T. K., Rijal, P., Coombes, N., Bewley, K. R., Tree, J. A., Radecke, J., Paterson, N. G., Supasa, P., Mongkolsapaya, J., Screaton, G. R., Carroll, M., Townsend, A., Fry, E. E., Owens, R. J. and Stuart, D. I. (2020). Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe 28(3): 445-454.e6.
Starr, T. N., Czudnochowski, N., Liu, Z., Zatta, F., Park, Y.-J., Addetia, A., Pinto, D., Beltramello, M., Hernandez, P., Greaney, A. J., Marzi, R., Glass, W. G., Zhang, I., Dingens, A. S., Bowen, J. E., Tortorici, M. A., Walls, A. C., Wojcechowskyj, J. A., De Marco, A., Rosen, L. E., Zhou, J., Montiel-Ruiz, M., Kaiser, H., Dillen, J. R., Tucker, H., Bassi, J., Silacci-Fregni, C., Housley, M. P., di Iulio, J., Lombardo, G., Agostini, M., Sprugasci, N., Culap, K., Jaconi, S., Meury, M., Dellota Jr, E., Abdelnabi, R., Foo, S.-Y. C., Cameroni, E., Stumpf, S., Croll, T. I., Nix, J. C., Havenar-Daughton, C., Piccoli, L., Benigni, F., Neyts, J., Telenti, A., Lempp, F. A., Pizzuto, M. S., Chodera, J. D., Hebner, C. M., Virgin, H. W., Whelan, S. P. J., Veesler, D., Corti, D., Bloom, J. D. and Snell, G. (2021). SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597(7874): 97-102.
ter Meulen, J., van den Brink, E. N., Poon, L. L., Marissen, W. E., Leung, C. S., Cox, F., Cheung, C. Y., Bakker, A. Q., Bogaards, J. A., van Deventer, E., Preiser, W., Doerr, H. W., Chow, V. T., de Kruif, J., Peiris, J. S. and Goudsmit, J. (2006). Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 3(7): e237.
Yuan, M., Huang, D., Lee, C.-C. D., Wu, N. C., Jackson, A. M., Zhu, X., Liu, H., Peng, L., Gils, M. J. v., Sanders, R. W., Burton, D. R., Reincke, S. M., Prüss, H., Kreye, J., Nemazee, D., Ward, A. B. and Wilson, I. A. (2021a). Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science 373(6556): 818-823.
Yuan, M., Liu, H., Wu, N. C. and Wilson, I. A. (2021b). Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem Biophys Res Commun 538: 192-203.
Yuan, M., Wu, N. C., Zhu, X., Lee, C.-C. D., So, R. T., Lv, H., Mok, C. K. and Wilson, I. A. (2020). A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368(6491): 630-633.
Yuan, M., Zhu, X., He, W. T., Zhou, P., Kaku, C. I., Capozzola, T., Zhu, C. Y., Yu, X., Liu, H., Yu, W., Hua, Y., Tien, H., Peng, L., Song, G., Cottrell, C. A., Schief, W. R., Nemazee, D., Walker, L. M., Andrabi, R., Burton, D. R. and Wilson, I. A. (2022). A broad and potent neutralization epitope in SARS-related coronaviruses. Proc Natl Acad Sci U S A 119(29): e2205784119.
- Yuan, M, Wu, N C, Yu, W and Wilson, I(2023). Expression and purification of CR3022 Fab and IgG. Bio-protocol Preprint. bio-protocol.org/prep2191.
- Yuan, M., Wu, N. C., Zhu, X., Lee, C. D., So, R. T. Y., Lv, H., Mok, C. K. P. and Wilson, I. A.(2020). A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV . Science 368(6491). DOI: 10.1126/science.abb7269
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
