Advanced Search
Expression and purification of A. halodurans Cas1, Cas2 and Cas4
Last updated date: Oct 18, 2022 Views: 610 Forks: 0
Expression and purification of A. halodurans Cas1, Cas2 and Cas4
Yukti Dhingra, Hayun Lee, Dipali G. Sashital
Abstract
Cas1, Cas2 and Cas4 are CRISPR associated adaptation proteins. This protocol describes the specific steps for purification of Alkalihalobacillus halodurans (previously Bacillus halodurans) Cas1, Cas2 and Cas4. These protocols can also be used for Cas1, Cas2, Cas4 and Cas4/1 fusion proteins from other organisms. The purified proteins can be used for biochemical assays and structural work to study the role of these proteins in CRISPR adaptation.
Background
CRISPR (Clustered regularly interspaced palindromic repeats) systems are adaptive immune systems that protect bacteria and archaea against foreign nucleic acids (reviewed in Jackson et al., 2017; Lee and Sashital, 2022). Short DNA sequences called spacers are acquired from invading DNA and stored within the CRISPR array as memories of infection, thus making the system adaptive. The CRISPR array is transcribed and processed to short CRISPR RNAs that are used to neutralizing the phage infection by RNA-mediated interference. Cas1, Cas2 and Cas4 form a complex and add new spacers to the CRISPR array. These proteins have been studied biochemically and structurally to understand the mechanisms of CRISPR adaptation. While purification of Cas1 and Cas2 is straightforward, purification of Cas4 can be challenging. Cas4 proteins contain iron sulfur clusters that do not always assemble properly. Also, the protein tends to precipitate during purification.
For all three proteins, we use Ni-NTA affinity chromatography as the first step of purification. For Cas4, this is followed by size exclusion chromatography. For Cas1 and Cas2 which are expressed with a maltose binding protein (MBP) solubility tag, we use tobacco etch virus (TEV) protease to cleave the tag. We then separate MBP from Cas1 or Cas2 using subtractive Ni-NTA chromatography and sometimes also, cation exchange chromatography using a heparin column. This is then followed by size exclusion chromatography.
We use the following constructs for expression of the proteins in E. coli:
Cas1/pSV272 – Kanamycin resistant - encodes His6-MBP-Cas1 under T7 promoter
Cas2/pSV272 - Kanamycin resistant - encodes His6-MBP-Cas2 under T7 promoter
Cas4/pET52b – Ampicillin resistant - encodes His6-Cas4 under T7 promoter
Cas4 is co-expressed with suf operon using sufABCDSE/pACYC construct (Chloramphenicol resistant) for better assembly of the iron-sulfur cluster domain of Cas4.
Materials Required:
| Reagent | Source | Catalog number |
| Escherichia coli NEB 5-alpha | New England Biolabs | C2987H |
| Escherichia coli Bl21 (DE3) | New England Biolabs | C2527H |
| Escherichia coli Bl21 (DE3) Overexpress C41 | Sigma | CMC0017 |
| HEPES | Thermo Fisher Scientific | BP3101 |
| Sodium phosphate dibasic heptahydrate | Thermo Fisher Scientific | S373-3 |
| Sodium Chloride | Thermo Fisher Scientific | S64010 |
| Imidazole | Thermo Fisher Scientific | O3196-500 |
| Potassium Chloride | RPI | P2173 |
| Glycerol | Thermo Fisher Scientific | G33-1 |
| DTT | Thermo Fisher Scientific | D11000-100.0 |
| PMSF | RPI | P20270-25.0 |
| Brilliant blue R-250 | RPI | B43000-50.0 |
| SDS | Thermo Fisher Scientific | 151-21-3 |
| Luria Broth | Thermo Fisher Scientific | BP14262 |
| Tryptone | RPI | 50213718 |
| Yeast Extract | Thermo Fisher Scientific | DF0127-07-1 |
| Ferrous sulfate | Fisher Scientific | I146-500 |
| Ferric citrate | Fisher Scientific | 3522-50-7 |
| Ammonium ferric citrate | Fisher Scientific | I72500 |
| L-cysteine free base | MP Biomedicals | 52-90-4 |
| IPTG | Fisher Scientific | BP1755100 |
| Kanamycin monosulfate | RPI | BP906-5 |
| Ampicillin | RPI | BP176025 |
| Chloramphenicol | RPI | BP904-100 |
| HisPur Ni-NTA resin | Thermo Fisher Scientific | 88223 |
| HiTrap Heparin HP | GE Healthcare | 17040703 |
| Superdex 200 Increase 10/300 | Cytiva | 28990944 |
| HiLoad 16/600 Superdex 200 | GE Healthcare | 28989335 |
| HiLoad 16/600 Superdex 75 | GE Healthcare | 28989333 |
Equipment
| 37°C/18°C Incubator shaker |
| Centrifuge with cool function |
| Branson Sonicator (Thomas Scientific SFX150) |
| Bio-rad NGC |
Procedure
Preparation for expression (about 2h)
Cas1 and Cas2:
- Prepare LB agar plates supplemented with 50 µg/mL of kanamycin.
- Prepare chemically or electrically competent Escherichia coli BL21(DE3) cells.
- Prepare 4 L of LB (1 L per 2.5 L flask) and 50 mL of LB (in a 250 mL flask). Autoclave using at least 45 min liquid cycle.
Cas4:
- Prepare LB agar plates supplemented with 100 µg/mL of ampicillin and 34 µg/mL of chloramphenicol.
- Prepare chemically or electrically competent Escherichia coli C41(DE3) cells.
- Prepare 4 L of 2XYT (1 L per 2.5 L flask) and 50 mL of 2XYT or LB (in a 250 mL flask). Autoclave using at least 45 min liquid cycle.
- Prepare stock solutions for ferrous sulphate, ferric citrate and ammonium ferric citrate- to be added at the time of induction. For ferrous sulphate and ammonium ferric citrate – 100 mg/mL stock (1000x) and for ferric citrate – 5 mg/mL (100x). Filter sterilize (0.22 μm) all the solutions and store the tubes/bottles covered with foil.
Preparation for purification (about 2h)
- Prepare 5x Ni-NTA buffer stock for Ni-NTA affinity purification buffers. The buffer compositions are listed in recipes section.
- Filter with a 0.22 µm vacuum filter.
- This can be prepared in larger volumes (1-2 L) and stored at room temperature.
- Prepare 500 mL of 3 M Imidazole, pH 8.0 for Ni-NTA affinity purification buffers.
Day 0 - Transformation (about 2 hours)
- Transform the following to the respective cells:
| Plasmids | Competent cells | Antibiotics |
| Cas1/pSV272 | Escherichia coli BL21(DE3) | 50 µg/mL Kanamycin |
| Cas2/pSV272 | Escherichia coli BL21(DE3) | 50 µg/mL Kanamycin |
| Cas4/pET52b + sufABCDSE/pACYC | Escherichia coli C41(DE3) | 100 µg/mL Ampicillin + 34 µg/mL Chloramphenicol |
Use ~100 ng of each plasmid (in a volume less than 10 µL) for a 100 µL aliquot of cells.
- After adding plasmids (and gently flicking tube) incubate on ice 30 min.
- Heat shock at 42°C for 30 s in a water bath and incubate on ice for 5 min.
- Add 900 µL of LB and incubate at 37°C for 1 h.
- Plate 100 µL of the recovery for Cas1 and Cas2 on LB plates with kanamycin.
- For Cas4, Centrifuge the cells at 4000 x g for 3 min, decant ~900 µL of media and resuspend cells in remaining liquid. Plate all the cells on LB plate with Ampicillin and Chloramphenicol.
- Incubate at 37 °C overnight.
Day 1 - Starter Culture (about 15 - 20 min)
1. Starting the culture
- Pick a single colony and grow in 50 mL LB (supplemented with appropriate antibiotics) overnight at 37°C.
- Loosely cover the flask with foil.
2. Prepare buffers for Ni-NTA column. These should be made fresh for each purification.
The following volumes should be sufficient:
- 100 mL lysis buffer
- 250 mL wash buffer
- 100 mL elution buffer
- Do not add DTT or PMSF to buffers until the day of the purification.
3. Prepare four 12% SDS-PAGE gels. Wrap gels in wet paper towels and plastic wrap. Store in 4 °C.
Day 2 –Expression
Cas1 or Cas2 (about 2 - 3 h)
- To each 1 L LB flask add 50 µg/mL kanamycin using 1 mL of 1000x stock.
- Inoculate each flask with 10 mL of the overnight starter culture.
- Grow cells to mid-log phase at 37 ºC with shaking at 180 rpm.
- When cells reach 0.4-0.5 OD600, add 1 mM ITPG using 1 mL of a 1 M IPTG stock.
- Reduce temperature to 16-18ºC and continue growth overnight with shaking at 180 rpm.
Cas4 (about 2 - 3 h)
- To each 1 L 2XYT flask add 100 µg/mL ampicillin and 34 µg/mL chloramphenicol using 1 mL of 1000x stocks.
- Inoculate each flask with 10 mL of the overnight starter culture.
- Grow cells to mid-log phase at 37ºC with shaking at 180 rpm.
- When cells reach 0.4-0.5 OD600, add 1 mM ITPG using 1 mL of a 1M IPTG stock.
- Add 100 mg solid L-cysteine to each flask (store L-cysteine in vacuum sealed desiccator).
- Add 1 mL of 1000x stocks of ferrous sulfate and ammonium ferric citrate and 10 mL of 100x stock of ferric citrate (final concentrations – 0.1 mg/mL, 0.1 mg/mL and 0.05 mg/mL respectively)
- Reduce temperature to 16-18ºC and continue growth overnight with shaking at 180 rpm.
Day 3: Purification step 1 (about 2h)
1. Harvest the cells by centrifuging at 4000 x g for 20 minutes.
- Weigh bottles before adding cultures and note their weights.
- After centrifugation, pour media into emptied flasks.
- Weigh the bottles containing the pellets and subtract initial bottle weight to determine the wet weight of the pellets.
Note: Typical pellets from LB and 2XYT growth should be ~5 g and ~9 g wet weight, respectively.
2. Resuspend the cells in lysis buffer.
- Add 100 µM PMSF and 2 mM DTT (DTT is necessary only for Cas4) to lysis buffer.
- Typically, 1 mL of buffer can be added per gram of pellet. You may need to add a bit more to fully resuspend the pellet, but it should not be more than 1.2x more (the cells should not be too dilute when you move on to sonication). The final volume of the resuspended cells should be slightly less than 2x the volume of lysis buffer you used.
- Store at -80 °C for future use or continue to the next step.
Day 3/4 - Purification step 2 (2 days for Cas1 or Cas2; 1 day for Cas4)
Ni-NTA affinity chromatography
- If cells were frozen, thaw in a water bath set in ice.
- Once thawed, add another 100 µM PMSF to cells (this is not necessary if continuing directly after resuspending cells).
- Transfer cells to a small plastic beaker.
- Lyse cells by sonication using a blunt tip sonicator.
- Use sonication settings: 15 s (0.55 s on 0.55 s off) at 80%.
- Rest for at least 30s between blasts. 3-4 blasts should be enough to lyse the cells.
Note: When fully lysed, lysate should change color (becoming slightly browner) and become very liquidy. It is a good idea to use a pipet to test the consistency of the lysate.
- Transfer lysate to 50 mL round bottom tubes and centrifuge at 20,000 x g for 30 min at 4 °C to clear cell debris.
- Spin a small amount (100 µL) of lysate separately in a 1.5 mL tube to prepare the pellet and lysate for gel analysis.
- Remove and collect the supernatant in a 1.5 mL tube.
- Resuspend the pellet in 100 µL lysis buffer.
- Prepare gel samples by diluting 2 µL of the supernatant (lysate) or resuspended pellet with 13 µL of lysis buffer and 5 µL 4x SDS-loading dye.
- While lysate is spinning, prepare a Ni-NTA column.
- The amount of resin will depend on the amount of lysate, but be careful not to use too much resin.
- For ~50 mL of lysate using 1 mL Ni-NTA resin should be sufficient. For this, 2 mL of resin slurry (50:50 resin:20% ethanol) can be added to a plastic column with a stopcock.
- After the liquid from the slurry flows through, wash the column with two full columns of ddH2O.
- Equilibrate the column with the remaining lysis buffer (at least two full columns).
- Load lysate carefully on the Ni-NTA column.
- The first few milliliters can be added using a pipet to ensure that the column bed does not get disrupted.
- Apply the lysate with the stopcock closed.
- Once all of the lysate is added (or the column is full), open the stopcock to start lysate flowing through the column.
- Collect the flow through in an empty beaker.
- If the total lysate volume does not initially fit in the column, continue to apply lysate as is flows through the column, being careful not to disturb the column bed.
- Do this until all lysate has flowed through the column.
- Prepare a sample of the flow through for gel analysis: 2 µL flow through, 13 µL lysis buffer, 5 µL 4X SDS loading dye.
- Add 2 mM DTT (DTT is necessary for only Cas4) to the wash buffer. Wash resin with wash buffer. Be careful not to disrupt the column when adding the buffer (the first several milliliters can be added with a pipet). Collect the wash in a fresh beaker. As you wash, you can check the absorbance of the eluate coming off the column on a spectrophotometer blanked with wash buffer. Wash the resin until A280 is ~0.1 and the A260/A280 ratio has reached ~1.3. Usually this will take ~50 column volumes. Once the absorbance has reached 0.1, let all of the wash buffer flow through, then proceed to the next step. Prepare a sample to analyze the wash fraction: 15 µL Wash, 5 µL 4X SDS loading dye.
- Add 2 mM DTT (DTT is necessary for only Cas4) to elution buffer. Close stopcock and carefully add several milliliters of elution buffer to the column (use a pipet for the first few milliliters). Open the stopcock to begin eluting the protein, collecting fractions that are approximately half a column volume (e.g. 500 µL fractions if using a 1 mL column).
Note: Fractions containing Cas4 will have a yellow brownish color. Prepare gel samples for each elution fraction: 15 µL fraction, 5 µL 4X SDS dye.
- Denature gel samples at 95ºC for 5 min then load 10 µL of each sample on 12% SDS PAGE gel. Stain the gel with Coomassie Brilliant Blue R250.
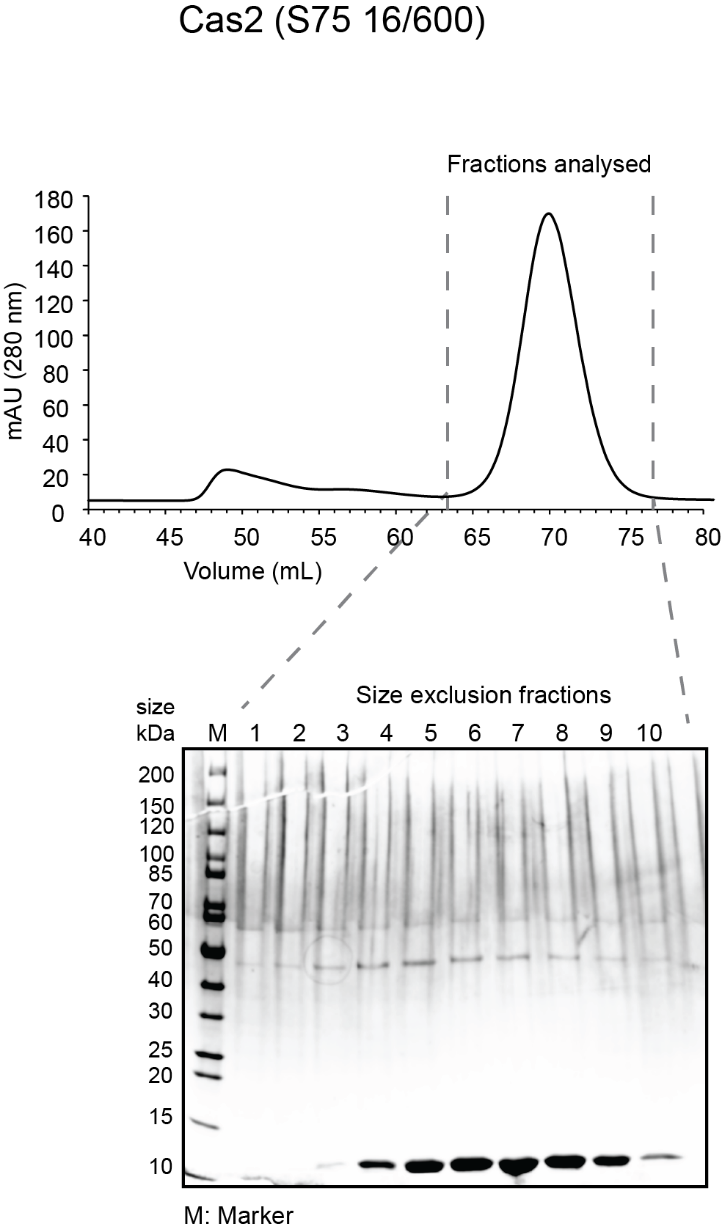
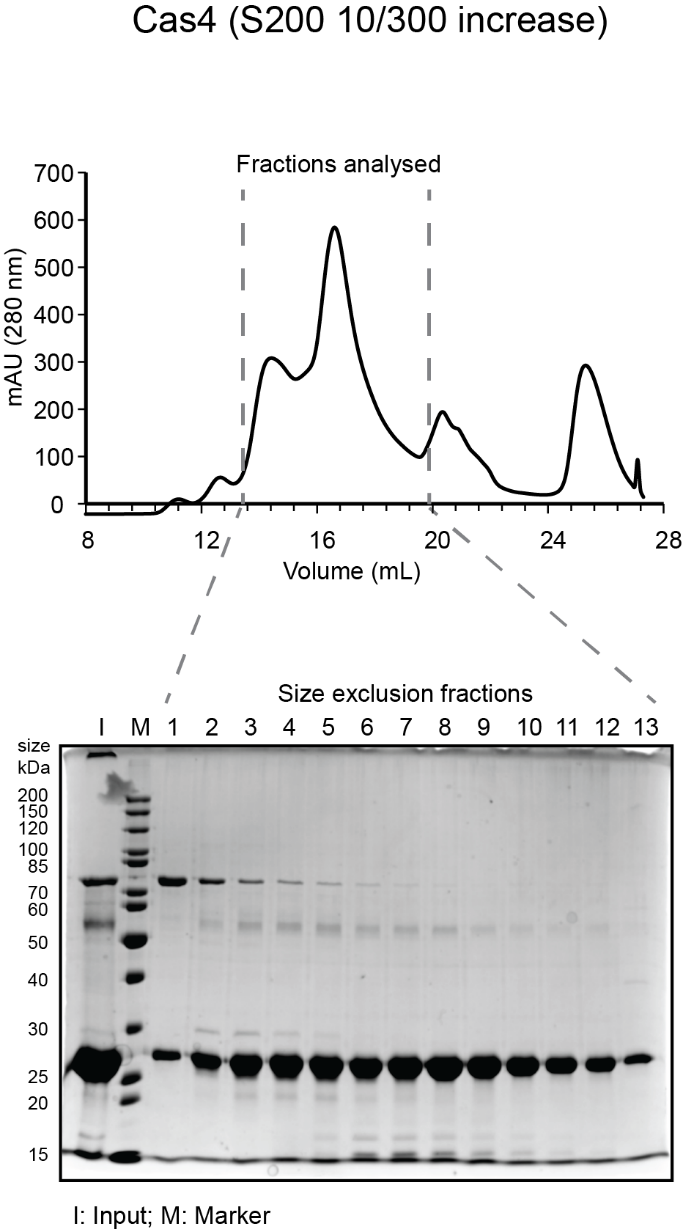
- While gel is running, analyze fractions on a spectrophotometer (blanked with elution buffer) to determine amount of protein in each fraction. Typically, the most concentrated fractions will have A280 of 2.0-4.0 and A260/A280 of 0.6-0.7. Expected sizes for His6-MBP-Cas1 and His6-MBP-Cas2 are 84 kDa and 55.5 kDa. Expected size for His6-Cas4 is 27 kDa. See sample gels below for Cas1 and Cas4:
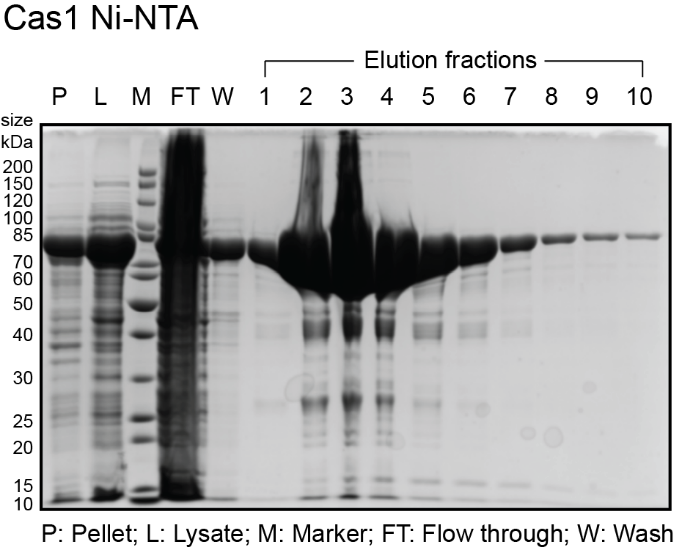
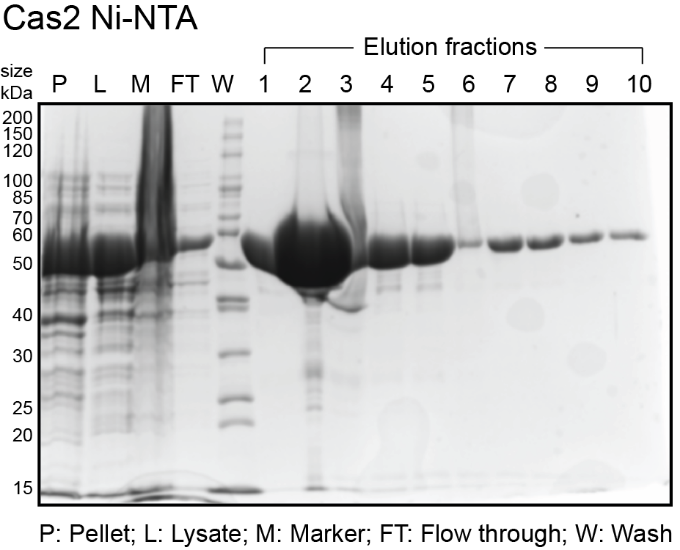
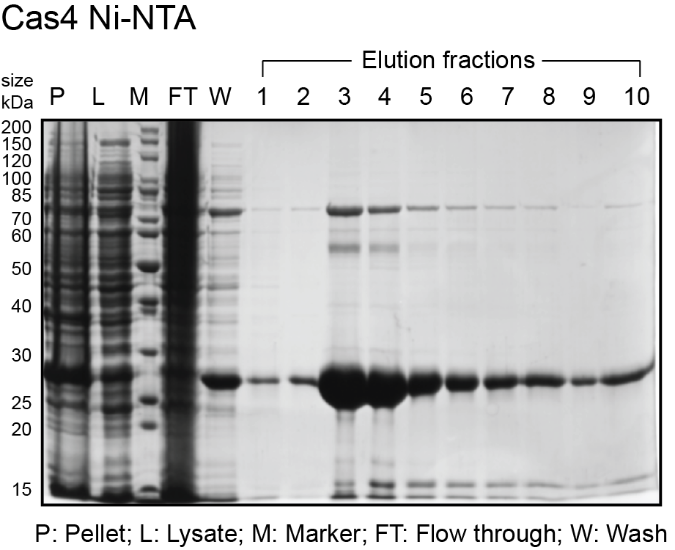
The protocol from here on is different for Cas4. Skip the Cas1 and Cas2 section to continue with Cas4.
Post Ni-NTA for Cas1 and Cas2:
- Pool only the most concentrated fractions in a new pre-chilled 50 mL tube. Mix the pooled fractions and measure concentration on a spectrophotometer with elution buffer as blank. Typically, the concentration of the pooled fraction is ~3 - 5 mg/mL. If the pooled fraction is highly concentrated, it may be useful to dilute the protein by adding elution buffer. Save 20 μL of this pooled fraction for analysis on gel the next day.
- Add TEV protease at 1:50 ratio (1 mg TEV for 50 mg of protein). We express and purify TEV in the lab and store it in 500 µL aliquots of at 0.5 mg/mL at -80 °C. (pRK793 https://www.addgene.org/8827/)
- Add DTT to dialysis buffer. Cut the required length of dialysis tubing (3.5 kDa MWCO) and soak it in dialysis buffer for 5 – 10 min. Transfer the pooled elution fractions + TEV into the dialysis bag and leave in the dialysis buffer overnight at 4 ºC with constant stirring.
Post Ni-NTA for Cas4
- Pool only the most concentrated fractions and concentrate to <1 mL using a 4 mL 10K MWCO concentrator. Use short spins of 10 min each at 4000 x g. At the end of each spin, resuspend the protein in the concentrator to redistribute the Cas4. You will likely see that the Cas4 is more concentrated at the bottom of the concentrator, based on the dark brown color in that area. This will prevent the protein from precipitating. After the protein is resuspended, add more of the pooled fractions and continue concentrating.
Skip the next section on Cas1 and Cas2 to continue with Cas4 purification.
Day4/5 - Cas1 and Cas2 - Purification step 3
Subtractive Ni-NTA
- Transfer dialyzed protein sample to a pre-chilled 50 mL tube.
- Subtractive Ni-NTA
- Equilibrate the Ni-NTA column with 3 CV dialysis buffer
- Apply the dialyzed protein sample to the column and collect flow through in two fractions.
- Wash the column with dialysis buffer and collect 5 fractions of 1 CV each
- Elute the remaining protein with 5 CV of elution buffer in 2 CV fractions
- Prepare gel samples for pre-TEV (from the previous day), post-TEV, flow through, wash and elution fractions 15 µl sample and 5µl of 4x SDS dye.
- Denature gel samples at 95ºC for 5 min then load 10 µL of each sample on 12% SDS PAGE gel. Typically, for MBP-Cas1, TEV activity and MBP cleavage is >80%. Expected sizes are 44.5 kDa for His6-MBP and 39.5 kDa for Cas1.
- If the flow through and wash fractions from subtractive Ni-NTA do not have any MBP, pool these fractions and concentrate them down to 1 mL volume for size exclusion chromatography using a 10 kDa MWCO concentrator and skip the next section for cation exchange chromatography. Sample gels for Cas1 and Cas2 are shown below. For this particular purification, cation exchange chromatography was skipped as the flow through was clean to proceed with size exclusion chromatography.
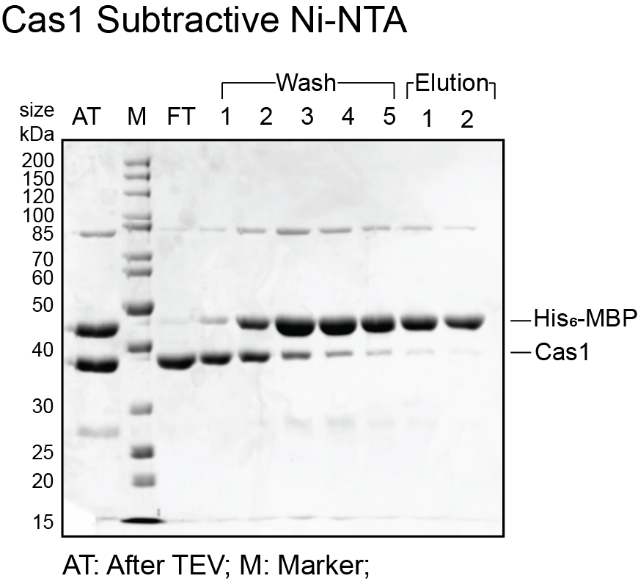
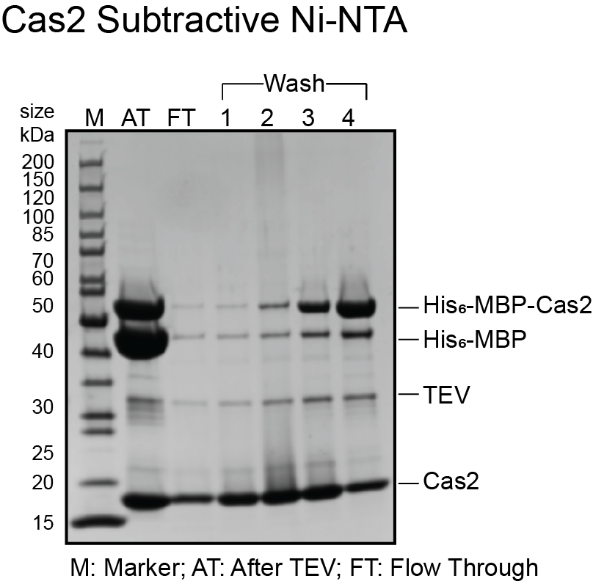
Cation exchange chromatography with HiTrap Heparin
- If the fractions have MBP, pool the flow through and wash for dialysis into a low salt buffer (Buffer A) for cation exchange chromatography using a HiTrap Heparin column. We use Bio-Rad NGC system to perform ion exchange and size exclusion chromatography.
- Switch the heparin column from ethanol to water at 2.5 mL/min (assuming the heparin column was stored in 20% ethanol). After the water wash, run high salt buffer (Buffer B) through the system (excluding the column). Then wash the system and equilibrate the heparin column with Buffer A (5 CV). Place 5 mL fraction collection tubes in the fraction collection rack.
- Load the dialyzed protein on the heparin column. Start the fraction collector, set to 5 mL fraction volume. Apply protein to the heparin column using a 5 mL sample loading loop at flow rate 5 mL/min. The chromatogram will show a rise in absorbance at 280 nm as the sample is being applied. When the absorbance drops, load the rest of the sample and repeat until all the sample has been applied. Wash the column with 10% Buffer B for 5 column volumes. Elute by applying a gradient from 10% to 100% Buffer B over a total volume of 25 ml.
- Prepare fractions from the gradient to run on a gel: 15 µL fraction, 5 µL 4X SDS loading dye. Run 10 µL of the sample on 12% SDS PAGE. Sample chromatograms and gels for Cas1 and Cas2 are shown below.
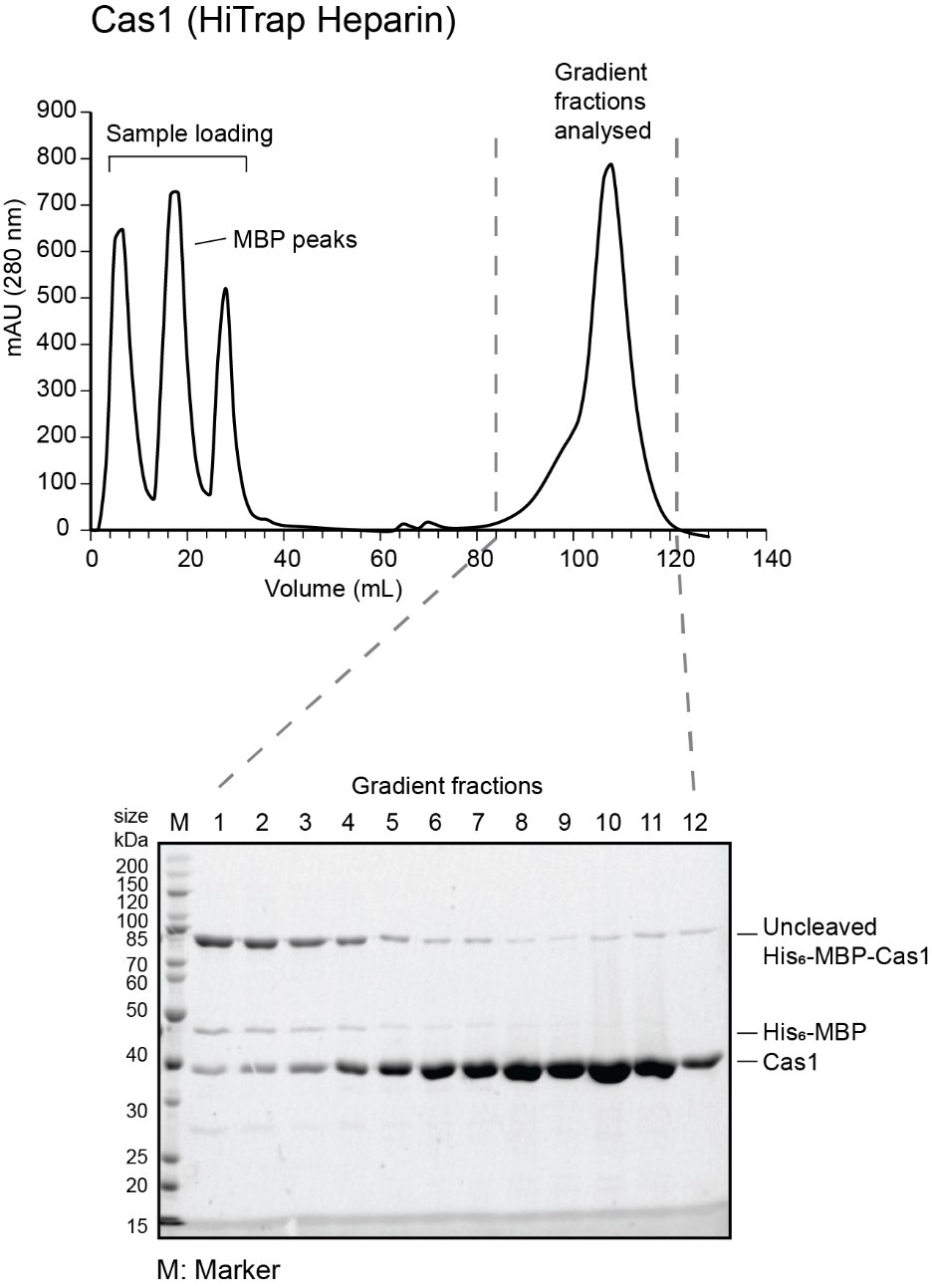
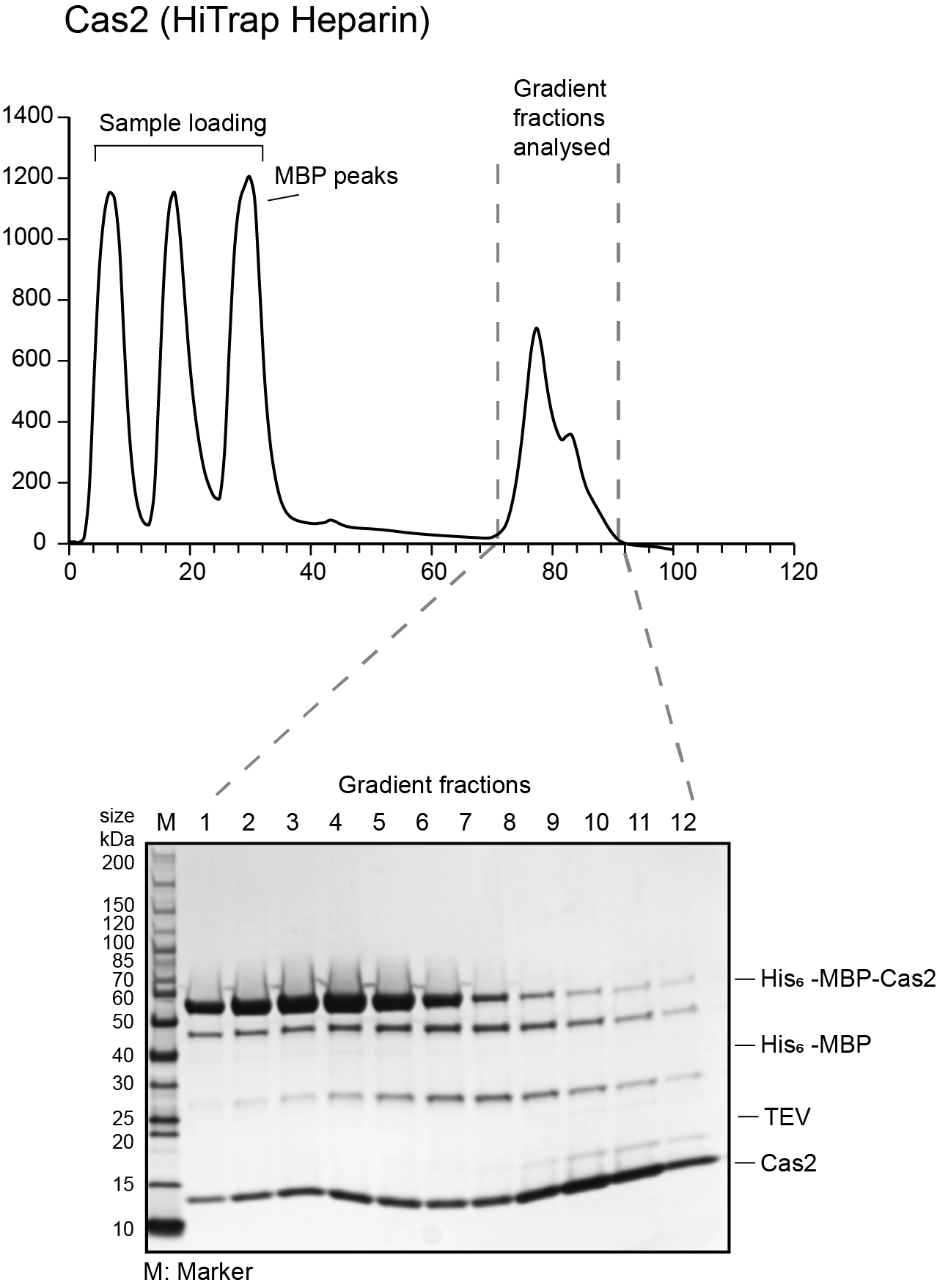
- Pool the cleanest fractions and concentrate them down to 1 mL volume for size exclusion chromatography using a 10 kDa MWCO concentrator. Use short spins of 10 min each at 4000 x g. At the end of each spin, resuspend the protein in the concentrator to redistribute the protein.
- Clean the heparin column and system with 100% buffer B, 10 CV of water and switch to 20% ethanol.
Day 3/4 (Cas4) Day 4/5 (Cas1 and Cas2) – Last purification step
Size exclusion chromatography
- Add DTT to SEC buffer. Equilibrate the S75 10/300 or S75 16/600 column for Cas1 or Cas2 with SEC buffer with 100 mM KCl. For Cas4, equilibrate S200 10/300 increase or S200 16/600 column with SEC buffer with 500 mM KCl. You can choose between the 10/300 or 16/600 columns based on how much protein you have and how much the column can be loaded on the column.
- Thoroughly rinse out the sample loading loop with water then buffer.
- For Cas4, in case of precipitation, spin at top speed at 4°C for at least 10 min (30 min is better). Carefully remove supernatant as soon as centrifuge has stopped using a 1 mL pipet, and transfer to a fresh tube. There may be a small pellet of precipitate, so be careful not to disturb it. Be conservative when removing the supernatant if there is a pellet, leaving behind ~10 µL of liquid.
- Apply protein to column and elute at the suggested flow rate (0.45 mL/min for 10/300 and 1 mL/min for 16/600). Collect 0.5 mL fractions for 10/300 column starting at 8 mL or 2 mL fractions for 16/600 column starting at 40 mL.
- Prepare fractions for gel: 15 µL fraction, 5 µL 4X SDS loading dye. Run 10 µL of input and size exclusion fractions on 12% SDS PAGE.
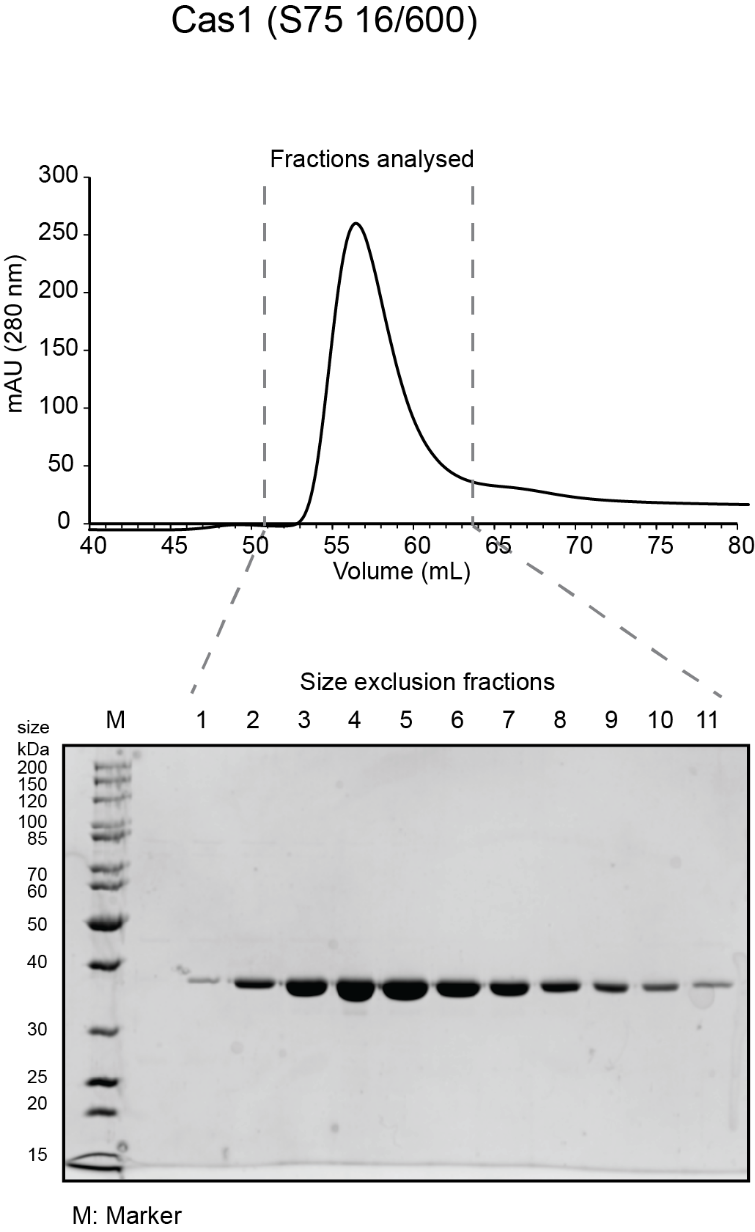
- Pool the peak fractions that are clean and have highest protein concentration. For the sample gels shown, fractions pooled were 4-9 for Cas1; 4-9 for Cas2 and 6-11 for Cas4. Concentrate (as before) to the desired concentration (~100 µM is ideal), aliquot in small volumes (5 µL -50 µL depending on intended use) in microcentrifuge tubes, flash freeze in liquid nitrogen and store at -80ºC.
- Sample chromatogram and gel for a Cas1 and Cas2 run on S75 16/600 and Cas4 run on S200 10/300 are shown below.
Recipes
| Ni-NTA column buffers | |||
5x Ni-NTA Buffer, pH 8.0 250 mM Sodium Phosphate Dibasic 2.5 M Sodium Chloride 25 % Glycerol | |||
Lysis buffer 1x Ni-NTA pH 8.0 Protease inhibitor (0.2 mM PMSF: add just before use) | Wash buffer 1x Ni-NTA pH 8.0
| Elution buffer 1x NiNTA pH 8.0
| |
| Dialysis and pre-Heparin prep buffers | |||
Dialysis Buffer (to be used alongside TEV cleavage) 1x Ni-NTA Buffer 2 mM DTT (add just before use) | |||
| Heparin column buffers | |||
Buffer A: 20 mM HEPES-KOH pH 7.5 100 mM KCl or 100 mM NaCl 5% Glycerol | Buffer B 20 mM HEPES-KOH pH 7.5 2 M KCl or 2 M NaCl 5% Glycerol | ||
| Size exclusion column buffer | |||
SEC buffer: 20 mM HEPES-KOH pH 7.5 5% glycerol 1mM DTT (add before use) | |||
| 100 mM PMSF (dissolve ~17 mg PMSF in 1 mL isopropanol just before use) | |||
Acknowledgements
This protocol was adapted from Lee et al., 2018, 2019 and Dhingra et al., 2022. This work was supported by an NIH R01 grant (GM115874) and NIH R35 grant (GM140876) to DGS. We thank all the members of the Sashital Lab for helpful input throughout the course of this project.
References
Jackson, S.A., McKenzie, R.E., Fagerlund, R.D., Kieper, S.N., Fineran, P.C., and Brouns, S.J.J. (2017). CRISPR-Cas: Adapting to change. Science (80-. ). 356. https://doi.org/10.1126/science.aal5056.
Lee, H., and Sashital, D.G. (2022). Creating memories: molecular mechanisms of CRISPR adaptation. Trends Biochem. Sci. https://doi.org/10.1016/j.tibs.2022.02.004.
Lee, H., Zhou, Y., Taylor, D.W., and Sashital, D.G. (2018). Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays. Mol. Cell 70, 48-59.e5. https://doi.org/10.1016/j.molcel.2018.03.003.
Lee, H., Dhingra, Y., and Sashital, D.G. (2019). The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. Elife 8. https://doi.org/10.7554/eLife.44248.
Dhingra, Y., Suresh, S.K., Juneja, P., Sashital, D.G. (2022). PAM binding ensures orientational integration during Cas4-Cas1-Cas2 mediated CRISPR adaptation . bioRxiv 1–41. https://doi.org/10.1101/2022.05.30.494039
Related files
 Bioprotocol_Cas_proteins_elife_Lee_2019.pdf
Bioprotocol_Cas_proteins_elife_Lee_2019.pdf - Dhingra, Y, Lee, H and Sashital, D G(2022). Expression and purification of A. halodurans Cas1, Cas2 and Cas4. Bio-protocol Preprint. bio-protocol.org/prep1996.
- Lee, H., Dhingra, Y. and Sashital, D. G.(2019). The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. eLife. DOI: 10.7554/eLife.44248
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
