Advanced Search
Flow cytometry–based ICAM1-binding assay
Last updated date: Sep 21, 2022 Views: 703 Forks: 0
The major T cell integrin, LFA1, is activated on T cell in response to chemokines and TCR crosslinking. During activation LFA1 clusters and increases in affinity, resulting in binding of ICAM1. This flow cytometry-based assay measures ICAM1-binding (both avidity and affinity) by fixing ICAM1-bound cells at desired timepoints following crosslinking of aCD3.
References: https://www.ncbi.nlm.nih.gov/pubmed/26740009 ; https://www.ncbi.nlm.nih.gov/pubmed/16458321 ; https://www.science.org/doi/10.1126/scisignal.abl9169
Special buffers and reagents:
- ICAM1-buffer: PBS + 0.5%BSA + 0.5mM Mg2+ + 0.9mM Ca2+ (Store in fridge for 1-2 months).
- Recombinant ICAM-1-Fc from R&D (cat: 796-IC-050).
- APC-anti-human Fcγ specific IgG F(ab’)2 from Jackson Immunoresearch (109-135-098) (can use PE-conjugated instead)
Methods:
For each time course of a given sample (for example genotype), 1x10e6 cells will be stimulated and aliquots taken out at different timepoints. This gives enough cells for 6 timepoints. We usually use 0 min, 1 min, 2 min, 5 min, 15 min, 30 min timepoints.
Preparation of T cells:
T cells are prepared for the assay as follows:
- If cells have been in culture, cells are rested for 2 hours without IL2 in media in 37˚C incubator.
- If inhibition of p110delta is desired, inhibition is done for 2 hours in 37˚C incubator prior to coating with aCD3.
- Cells are coated with 1 µg/ml anti-CD3e (2C11) (1x10e6 cells/ml) on ice for 30 min in ICAM1-buffer. 1x10e6 cells gives ≈160.000 cells/timepoint with 6 timepoints. This coating is done in 1.5 mL Eppendorf tubes. – Prepare scICAM1 during incubation.
Preparation of scICAM1:
- Prepare ICAM1-Fc-F(ab’)2 complexes in PBS – Mix per reaction (i.e. per sample):
• 7.9 µL PBS
• 1.3 µL APC- or PE-labelled anti-human Fcγ-specific IgG F(ab’)2 fragments
• 0.8 µL ICAM1-Fc (400 µg/ml stock) - Mix and leave on ice for 30 min or more. Prepare fresh before assay (I normally start the conjugation during the incubation of cells with aCD3). I make enough for the number of samples plus 15 % extra.
Preparation for assay:
- Prepare a strip of 8-strip tubes with 500 µL of 4 % PFA for each timepoint (generally 6/sample). Lower volumes than 500 µL (for example 250 µL) have also been used with success.
- Heat water bath or PCR machine to 37˚C (Make sure it’s 37˚C by thermometer).
- Prepare 12.5 µL/sample anti-Armenian hamster IgG in ICAM1-buffer at 20 µg/ml (4X final concentration; final concentration 5 µg/ml). We normally prepare 30 % extra.
- Prepare timer for timepoints, set pipettes for the different volumes needed for activating the cells (and if using a multichannel pipette prepare samples, scICAM1 and anti-armenian hamster IgG in a PCR plate so that it is ready to quickly add the volumes, see step 19).
Assay-time:
- Wash cells with ICAM1-buffer (2x with 0.5 mL ICAM1-buffer).
- Resuspend cells at 40µL/1x10e6 cells of ICAM1-buffer – keep on ice.
- The cell suspension volume is normally slightly higher than 40 µL as there is a residual volume from the wash step. 45 µL of cells are now transferred to an eppendorf tube or row A of the PCR plate prepared in step 9.
- Add 8 µL of ice-cold scICAM1 (prepared above).
- Take out 10 µL (can be done with multi-channel pipette) and add to 500 µL, 4 % PFA (prepared above) – this is the 0 min timepoint.
- Add 12.5 µL of prepared anti-armenian hamster IgG (final concentration 5 µg/ml) and quickly place in preheated waterbath or PCR machine – start timer.
- After desired timepoints, take out 10 µL and add to 500 µL, 4 % PFA (prepared above); this gives enough for 5 timepoints (as well as 0 min timepoint in step 14) – We usually use 1 min, 2 min, 5 min, 15 min, 30 min timepoints.
- After fixation, incubate all samples 5 min at RT.
- Spin down, and wash with FACS-buffer.
Optional: prepare PCR plates for multichannel pipettes so that multiple samples can be done simultaneously.
- Prepare the anti-Armenian hamster IgG and ICAM1-Fc-F(ab’)2 complexes in a PCR plate so that you can easily add the ICAM1-Fc-F(ab’)2 complexes and anti-Armenian Hamster IgG with a multichannel pipette to the samples.
- One row with samples (row A; See step 12)
- One row with ICAM1-Fc-F(ab’)2 complexes (row C; 9 µL/well; from step 4-5)
- One row with anti-Armenian hamster IgG (row D; 16 µL/well, from step 8)
Optional: Post assay
If cells are a mix of CD4+ and CD8+ T cells, CD4 and CD8 can be stained following fixation, however antibody concentrations should be titrated for fixed cells. CD8 staining in particular is often quite dim following fixation, so use a bright fluorophore such as BV421 or PE.
Analysis:
Samples are read on a flow-cytometer with a platereader and gated in the desired subset of analysis, and then gated for ICAM1+ and negative cells. See example of staining below (Fig. S1 from Johansen et al, 2022).
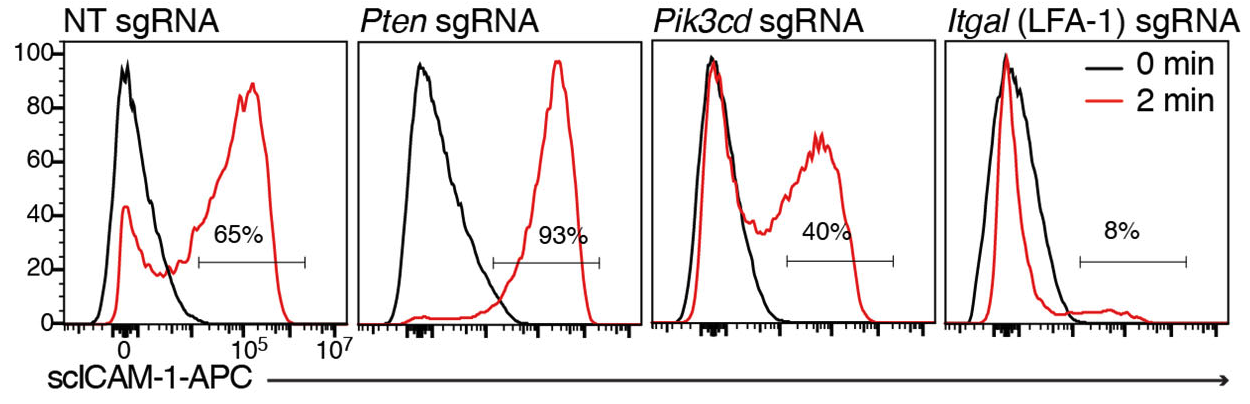
Gating is done based on 0 min timepoint, which should have little or no ICAM1-binding in WT cells. One can analyse ICAM1-binding either as MFI or as percentage of cells that bind ICAM1. In most cases these two methods of analysis give very similar trends, however if binding is very high (such as for Mn2+/Mg2+ stimulation), MFI analysis may be preferred.
Optional controls:
If desired, one can also stimulate with the following:
- PMA (50 ng/ml) – inside-out signalling
- Mg2+/EGTA (5-10mM/1mM) – outside-in signalling
- CCL19/SDF1/CCL21 (250 ng/ml) – chemokine response (earlier timepoints (30, 60, 120 sec)
For LFA-1 affinity experiments T cells are treated the same way as for the ICAM-1 complexes-based adhesion assay but incubated with monomeric ICAM-1 Fc instead of scICAM-1. After fixation the cells are stained for anti-CD4 and anti-human Fcγ.
- Johansen, K, Okkenhaug, K and Schwartzberg, P(2022). Flow cytometry–based ICAM1-binding assay. Bio-protocol Preprint. bio-protocol.org/prep1958.
- Johansen, K. H., Golec, D. P., Huang, B., Park, C., Thomsen, J. H., Preite, S., Cannons, J. L., Garçon, F., Schrom, E. C., Courrèges, C. J. F., Veres, T. Z., Harrison, J., Nus, M., Phelan, J. D., Bergmeier, W., Kehrl, J. H., Okkenhaug, K. and Schwartzberg, P. L.(2022). A CRISPR screen targeting PI3K effectors identifies RASA3 as a negative regulator of LFA-1–mediated adhesion in T cells. Science Signaling 15(743). DOI: 10.1126/scisignal.abl9169
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
