Advanced Search
Cell-to-cell fusion assay
Last updated date: Jul 13, 2022 Views: 1223 Forks: 0
Abstract
SARS-CoV-2 spike protein mediates the interaction with Ace2 and facilitates the viral entry process. Accordingly, mutations in the spike protein change its affinity with the hACE2 protein, increase fusogenicity, and can modulate sensitivity to neutralizing antibodies. Cells expressing spike from the Delta variant have a higher propensity to form syncytia with hACE2 expressing cells. Here, we describe an assay to examine the ability of SARS-CoV-2 spikeprotein to generate frequent multinucleated syncytia.
Keywords: SARS-CoV-2 virus, Spike protein, ACE2 protein, Cell-to-Cell fusion,
Materials and Reagents
- HEK293T cells (EACC)
- Plasmids encoding hACE2, SARS-CoV-2 spike protein, pEGFP-N1 (or any GFP encoding plasmid), pTagRFP657 (or any plasmid encodingRFP).
- DMEM (Biowest, USA Catalog number: L0102)
- Penicillin-streptomycin (Gibco, USA Catalog number: 15140-122)
- L-Glutamine (Gibco USA Catalog number: 25030-081)
- 24 well cell culture plate (Eppendorf)
- 96 well cell culture plate (Eppendorf)
- Trypsin-EDTA (GibcoTM, Catalog number: 25200-056)
- Phosphate buffer saline (PBS) (HyClone, USA Catalog number: SH30256.02)
- Hemocytometer
- CO2 incubator
- Fetal Bovine Serum (FBS) (Gibco, USA 10082-147)
- Inverted microscope with fluorescence imaging capabilities
- Hoechst 33342 (Sigma Aldrich, 14540-100G)
- Lipofectamine 3000 (Invitrogen, Catalog number: L3000008)
- Opti-MEMTM (Gibco, USA Catalog number: 31985070)1
Equipment and software
- An inverted fluorescence microscope that has channels for visualizing GFP and TagRFP657. The microscope should provide magnification up to 40X for better visualization.
- Image acquisition software for instrument control and image acquisition.
- ImageJ software for image processing and area measurement.
- Graphpad Prism for data analysis (any data analysis software can be used).
Procedure
- Seed HEK293T cells in 24 well plates at 0.2x106/ 0.5ml in complete DMEM medium and incubate in a CO2 incubator for 24 h. The plate format can be selected according to the requirement.
- After 24 h of seeding, transfect the cells with pEGFP-N1 (50 ng) and Ace2 expressing vectors(500ng), in anotherwell, co-transfect cells with pTag-RFP657 (50ng) and spikeΔ19 expressing vectors (500ng) using Lipofectamine 3000 protocol. Scalethe plasmid amounts used according to the plate format.
Transfection
Prepare DNA mixture and Lipofectamine in separate tubes.
a. DNA-mixture:
Use 500 ng of pcDNA 3.1(-) spike ∆19 and 50 ng of pTag-RFP657 or use pEGFP-N1 (50 ng) and ACE2 expressing vectors (500ng) plasmids and dilute the DNA mixture in 50 µl of Opti-MEM and add 1µl of P3000 reagents and mix well.
b. Lipofectamine-mixture:
Use 1 µl of Lipofectamine 3000 reagent in 50 µl of Opti-MEM per well and Incubate the Lipofectamine mixture for 5 minutes at room temperature.
c. Add DNA mixture on top of Lipofectamine mixture dropwise, gently tap the solution and incubate for 15 min at the room.
d. Add the transfection solution on top of the cells dropwise.
e. Incubate cells at 37 ∘C and %5 CO2 for 6 h.
f. After 6 h of transfection, removethe medium and replace it with a fresh complete DMEM medium (supplemented with 10%FBS and Glutamine). - After 10-12 h of transfection, wash the cells with 1x PBS (100 µl).
- Trypsinize the cells in both the wells using 50 µl of trypsin for 2 minutes at 37 ∘C.
- Add complete DMEM medium to the cells (500 µl/well) to neutralize trypsin and make single-cell suspension by pipetting.
- Next, take 5000 cells from each well and mix at a 1:1 ratio (ACE2: Spike) in 96 well plates containing 100 µl of complete DMEM medium.
- (Day 4) After 48-50 h of transfection, counter-stain the cells with Hoechst (1:20,000 final dilution) for 20 minutes at room temperature.
- Following Hoechst staining, wash the cells thrice with 1xPBS and fix using 4% paraformaldehyde (PFA) for 20 min at room temperature.
- To remove (PFA), wash the cells thrice with PBS and take them for imaging using the fluorescence microscope (Figure 1).
- To quantifythe area of syncytia formationby cell-to-cell fusion(GFP and RFP overlapping cells) quantify using ImageJ software.
Quantification of the area of syncytia
- Import the multi-channel images in the ImageJ.
- Identify the cells with multiple nuclei (in the blue channel) expressing both GFP and RFP.
- Select either the red or green channel (based on contrast and signal intensity. Higher intensity and contrast provide a good S/N ratio) for quantification of area.
- Convert the image into a binary image. This will convert the image into black and white (high contrast). It can be done manually by adjusting the threshold.
- Select the ‘Magic wand’ tool on the ImageJ toolbar.
- Click/select the previously identified syncytia (in step 12). The complete cell area will automatically be selected.
- For measuring the area, click on the ‘Analyze’ tab on ImageJand select measure.This will
calculate the number of pixels present in the selection. - Measure the area (in pixels) of ‘n’ number of syncytia for statistical analysis.
- Export the measurements for analysis.
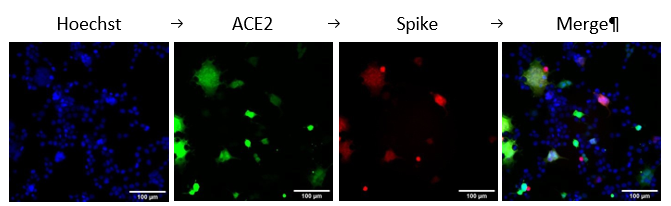
Figure 1. HEK293T ACE2 (green) and Spike (Red) cells were mixed, and syncytium formation was observed after 48h of transfection.
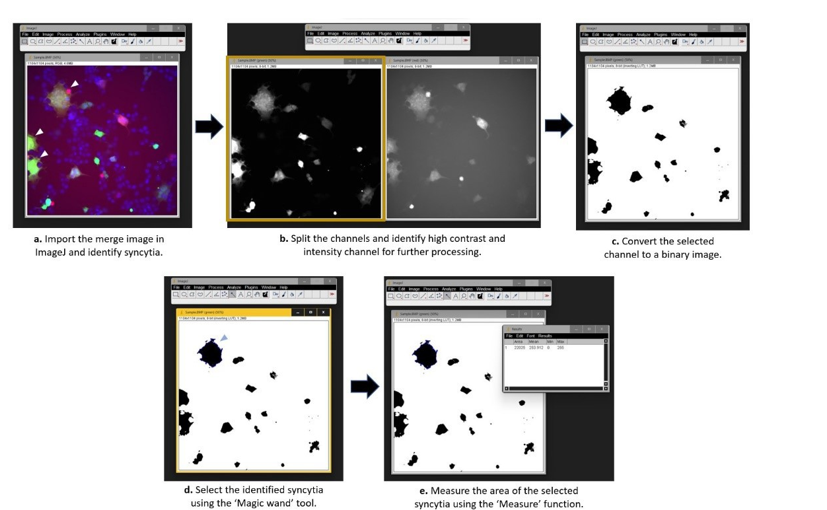
Figure 2. Stepwise instructions for quantification of area of syncytia.
Controls
Positive and Negative controls for the analysis are very important. You can use the spike from Wuhan or UK strain in addition to cells transfected only with RFP as control.
Troubleshooting
- If fusion is not observed.
# Note: make sure spike and hACE2 proteins are expressing.
# Do not disturb the plate after seeding cells in 96 well plate format. # Make sure the cells are free of any microbial contaminations. - The area measured is not correct
By using the ‘make binary’ option (Process < Binary < Make Binary), the software automatically sets a threshold, which may exclude the low signal obtained from the cell periphery leading to abnormal cell shape. Here we may lose some area from the syncytia. Another possibility is that two cells were close to each other and detected as one, so make sure that no two cells are joined. The ‘magic wand’ tool detects them as one cell. To troubleshoot this, adjust the threshold manually (Image < Adjust < Threshold) so that the low signal areas are also included. For cells that are very close to each other and detected as one, the ‘watershed’ option (Process < Binary < watershed) can be used to differentiate merged cells into individual cells.
- Mishra, T, Dalavi, R and Chande, A(2022). Cell-to-cell fusion assay. Bio-protocol Preprint. bio-protocol.org/prep1792.
- Mishra, T., Dalavi, R., Joshi, G., Kumar, A., Pandey, P., Shukla, S., Mishra, R. K. and Chande, A.(2022). SARS-CoV-2 spike E156G/Δ157-158 mutations contribute to increased infectivity and immune escape. Life Science Alliance 5(7). DOI: 10.26508/lsa.202201415
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
