Advanced Search
Urine-based Enzyme-Linked Immunosorbent Assay to detect anti-SARS-CoV-2 nucleocapsid protein antibodies
Last updated date: Jun 14, 2022 Views: 786 Forks: 0
Abstract
An in house urine-based ELISA (Enzyme-linked immunosorbent assay) were developed to detect anti-SARS-CoV-2 nucleocapsid protein (N) IgG antibodies in urine samples, providing a noninvasive diagnostic tool to assess the humoral response against COVID-19. The presence of anti-SARS-CoV-2 N protein antibodies was detected with 94% sensitivity and 100% specificity with urines collected from the second to the 60th day post symptoms onset with this protocol.
Background
Many different serological assays are available for COVID-19, which preferentially uses plasma or serum as sample. Drawing blood can be unpleasant and difficult to perform in some circumstances, requiring sterile equipment and trained staff. Body fluids such as urine have been suggested as an alternative for the detection of immunoglobulins against several microbial agents (1-10). In this context, we develop an in-house ELISA to detect anti- SARS-CoV-2 nucleocapsid protein antibodies in urine samples. The protocol was based on a pre-validated serum-based ELISA with some modifications. Urine-based and paired serum-based ELISA achieved a very similar qualitative profile. Some of the advantages of the urine-based ELISA include ease of sample collection, biological sample stability and high levels of accuracy. Since most vaccine platforms are based on immune response against SARS-CoV-2 Spike (S) protein, anti-SARS-CoV-2 N ELISA may differentiate in this case individuals previously infected with SARS- CoV-2 from those vaccinated (11,12). So, this test may contribute to epidemiological studies by helping to determine previous exposure to SARS-CoV-2 in a population or individual level.
Materials and Reagents
- High binding 96-well polystyrene microplate, (Corning, Merck, Germany, catalog number: CLS2592).
- SARS-CoV-2 Nucleocapsid Recombinant protein (in house production). *1
*1 Note: alternative N antigen from Fapon Biotech Inc, China, catalog number: FP0516. - Coating buffer: 0.2 M sodium carbonate/bicarbonate, pH 9.6 (Merck, Germany, catalog number: 222321 and S6014).
- Wash buffer: 0.1 M sodium phosphate (Merck, Germany, catalog number: 7558-79-4), 0.15 M sodium chloride (Merck, Germany, catalog number): S9625, pH 7.4 containing 1% Tween 20 (Merck, Germany, catalog number: P2287), pH 7.4.
- Blocking buffer: 1% (w/v) Bovine Serum Albumin (BSA, Merck, Germany, catalog number: A7030) in wash buffer pH 7.4.
- Conjugated antibody: peroxidase-conjugated anti-human IgG antibody (Sigma- Aldrich, A0170) diluted (1:10,000) in wash buffer.
- Substrate: TMB substrate (Scienco, Brazil, Reference: One Step).
- Stop solution: 0.5M sulfuric acid (Merck, Germany, catalog number: 258105).
- 50 mL reagent reservoir sterile polystyrene (Merck, Germany, catalog number: CLS4870).
- Tips: 10 µl, 200 µl and 300 µl.
Equipment
- Vortex Mixer (Thermo Fisher Scientific, USA, catalog number: 88882012).
- Incubator at 37°C (Fanem, Brazil, catalog number: 502/2-C).
- ELISA Microplate Washer (Wellwash™ Microplate Washer, 5165000).
- Microplate Spectrophotometer (Multiskan Go, Thermo Fisher Scientific, USA, catalog number: N10588, 450 nm).
- Microplate Washer (not compulsory, Wellwash Versa, Thermo Fisher Scientific, USA, catalog number: 5165010).
- Monochannel 1-10 µl pipette (Thermo Fisher Scientific, USA, catalog number: 4641040N).
- Monochannel 10-100 µl pipette (Thermo Fisher Scientific, USA, catalog number: 4641070N).
- Multichannel 30-300 µl pipette (Thermo Fisher Scientific, USA, catalog number: 4661070N).
Software
- Excel (Microsoft).
- GraphPad Prism (version 8.0,USA).
Procedure
1. Plates
- Coating: Distribute 100 µl of coating solution containing the antigen in each well of the plate (400 ng/well). Incubate at 2-8°C overnight;
Blocking: Remove the coating solution from the wells by aspiration (ELISA microplate washer) or by decanting (manual). Add 250 µl of blocking solution to each well of the plate. Incubate at room temperature for two hours;
- Drying: Remove the blocking solution, tap the plate inverted for a few seconds on absorbent paper until dry.
2. ELISA
- Place all reagents at room temperature and wait for 30 minutes until stabilize;
- Add 100 μL of each sample (undiluted urine) to the wells of the plate;
- Cover the wells with plate sealer;
- Incubate the plate for 60 minutes at 37°C;
- Remove the plate sealer;
- Discard the contents of the cavities by suction (washing machine) or by decanting (manual);
Add 300ul of Wash Solution to each well (and then remove) and repeat it for 5 times. To ensure plate drying, at the end of the wash, tap the plate inverted for a few seconds on absorbent paper;
- Add 100 μL of Conjugated antibody into each well;
- Cover the wells with plate sealer;
- Incubate the plate 60 minutes at 37°C;
- Remove the plate sealer;
- Repeat the Wash step (step g);
- Add 100 μL of Substrate (TMB) into all wells;
- Protect the plate from light;
- Incubate the plate at room temperature until the desired color intensity is reached; *2
*2 As established by the manufacturer, for Scienco TMB we used 30 minutes. - Add 100 µl of the Stop Solution into all wells;
- Measure the absorbance at 450nm.
3. Data analysis
a. Calculation of ELISA cut-off value.
The optical density (OD) of the negative control samples *3 was determined and used to calculate the Cut-off as determined by the formula (13):
*3 We used n=30 negative samples.
𝑐𝑢𝑡 − 𝑜𝑓𝑓 = 𝐴𝑛𝑒𝑔 + 2 (𝑆𝐷𝑛𝑒𝑔)
𝐴𝑛𝑒𝑔: Average of negative samples
𝑆𝐷𝑛𝑒𝑔: standard deviation of negative samples
Use the cut-off to evaluate the diagnostic performance by estimation of sensitivity, specificity, area under the curve at GraphPad Prism (Analyses>Column analyses>Roc Curve).
b. Index calculation: calculate the index (I) of sample absorbance (Abs) over the value of cut-off, according to the equation:
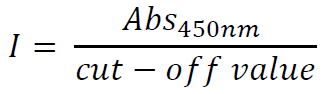
The results can be classified as follows:
I < 0.8: negative
0.8 ≤ I < 1.1: borderline (indeterminate)
I ≥ 1.1: positive
Use conditional formatting in Excel to classify the results.
In the case of an indeterminate result, the sample must be reanalysed. Samples that repeatedly obtain indeterminate results should be retested using an alternative method. If results remain indeterminate, a new sample should be collected in one week.
Acknowledgments
We acknowledge the help of all students, nurses, and clinicians involved in the collection and organization of the samples. We also thank Hospital das Clínicas/UFMG and Hospital Santa Helena for allowing the collection of samples. Last, we thank CT-Vacinas/UFMG for technical support. The study was supported with the following grants: Secretaria de Educação Superior do Ministério da Educação (SESU/MEC) - Grant 04/2020 - Enfrentamento da COVID-19; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) - Grant APQ-408675/2018-7; Brazilian Ministry of Science, Technology and Innovation (MCTI) - Rede Virus thought its many iniciatives ; Brazilian agencies Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), CNPq and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) - fellowships/scholarships.
Reference
C. Eamudomkarn, P. Sithithaworn, C. Kamamia, A. Yakovleva, J. Sithithaworn, S. Kaewke, Techasen, W. Loilome, P. Yongvanit, C. Wangboon, P. Saichua, M. Itoh, J. M. Bethony, Diagnostic performance of urinary IgG antibody detection: A novel approach for population screening ofstrongyloidiasis. PLOS ONE 13, e0192598 (2018).
- K. Sahni, A. Nagendra, P. Roy, S. Patrikar, Usefulness of enzyme immunoassay (EIA) for screening of anti HIV antibodies in urinary specimens: A comparative analysis. Med. J. Armed Forces India 70, 211–214 (2014).
- SS. Vázquez, S. Cabezas, A. B. Pérez, M. Pupo, D. Ruiz, N. Calzada, L. Bernardo, O. Castro, D. González, T. Serrano, A. Sanchez, M. G. Guzmán, Kinetics of antibodies in sera, saliva, and urine samples from adult patients with primary or secondary dengue 3 virus infections. Int. J. Infect. Dis. 11, 256–262 (2017).
- S. A. Ejazi, P. Bhattacharya, M. A. Bakhteya, A. A. Mumtaz, K. Pandey, V. N. Das, P. Das, M. Rahaman, R. P. Goswami, N. Ali, Noninvasive diagnosis of visceral leishmaniasis: Development and evaluation of two urine-based immunoassays for detection of Leishmania donovani infection in India. PLOS Negl. Trop. Dis. 10, e0005035 (2016).
- M. S. Joshi, S. D. Chitambar, V. A. Arankall, M. S. Chadha, Evaluation of urine as a clinical specimen for diagnosis of hepatitis A. Clin. Diagn. Lab. Immunol. 9, 840–845 (2002).
- M. Itoh, N. Ohta, T. Kanazawa, Y. Nakajima, M. Sho, M. Minai, Z. Daren, Y. Chen, H. He, Y. K. He, Z. Zhong, Sensitive enzyme-linked immunosorbent assay with urine samples: A tool forsurveillance ofschistosomiasis japonica. Southeast Asian J. Trop. Med. Public Health 34, 469–472 (2003).
X. G. Qiu, F. Nakamura-Uchiyama, Y. Nawa, M. Itoh, A tool for mass-screening of paragonimiasis: Anenzyme-linked immunosorbent assay withurine samples. Trop. Med. Health. 44, 19 (2016).
Y. Gong, Q. Li, Y. Yuan, Accuracy of testing for anti-Helicobacter pylori IgG in urine forH. pylori infection diagnosis: A systematic review and meta-analysis. BMJ Open 7, e013248 (2017).
S. Asfaram, S. Hosseini Teshnizi, M. Fakhar, E. S. Banimostafavi, M. Soosaraei, Is urine a reliable clinical sample for the diagnosis of human visceral leishmaniasis? A systematic review and meta-analysis. Parasitol. Int. 67, 575–583 (2018).
- S. A. Ejazi, A. Bhattacharyya, S. T. Choudhury, S. Ghosh, A. Sabur, K. Pandey, V. N. R. Das, P. Das, M. Rahaman, R. P. Goswami, N. Ali, Immunoproteomic identification and characterization of Leishmania membrane proteins as non-invasive diagnostic candidates for clinical visceral leishmaniasis. Sci. Rep. 8, 12110 (2018)
Bagno, F.F.; Andrade, L.A.F.; Sérgio, S.A.R.; Parise, P.L.; Toledo-Teixeira, D.A.; Gazzinelli, R.T.; Fernandes, A.P.S.M.; Teixeira, S.M.R.; Granja, F.; Proença-Módena, J.L.; da Fonseca, F.G. Previous Infection with SARS-CoV-2 Correlates with Increased Protective Humoral Responses after a Single Dose of an Inactivated COVID-19 Vaccine. Viruses 2022, 14, 510. https://doi.org/10.3390/v14030510
García-Montero, C.; Fraile-Martínez, O.; Bravo, C.; Torres-Carranza, D.; Sanchez-Trujillo, L.; Gómez-Lahoz, A.M.; Guijarro, L.G.; García-Honduvilla, N.; Asúnsolo, A.; Bujan, J.; et al. An Updated Review of SARS-CoV-2 Vaccines and the Importance of Effective Vaccination Programs in Pandemic Times. Vaccines 2021, 9, 433. [Google Scholar] [CrossRef]
Findlay JW, Smith WC, Lee JW, Nordblom GD, Das I, DeSilva BS, Khan MN, Bowsher RR. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective. J Pharm Biomed Anal. 2000 Jan;21(6):1249-73. doi: 10.1016/s0731-7085(99)00244-7. PMID: 10708409.
- Ludolf, F, Ramos, F F, Bagno, F, Coelho, E A and Fonseca, F G(2022). Urine-based Enzyme-Linked Immunosorbent Assay to detect anti-SARS-CoV-2 nucleocapsid protein antibodies. Bio-protocol Preprint. bio-protocol.org/prep1720.
- Ludolf, F., Ramos, F. F., Bagno, F. F., Oliveira-da-Silva, J. A., Reis, T. A. R., Christodoulides, M., Vassallo, P. F., Ravetti, C. G., Nobre, V., da Fonseca, F. G. and Coelho, E. A. F.(2022). Detecting anti–SARS-CoV-2 antibodies in urine samples: A noninvasive and sensitive way to assay COVID-19 immune conversion. Science Advances 8(19). DOI: 10.1126/sciadv.abn7424
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

