Advanced Search
Negative staining of HIV viral particles permeabilised with PFO with capsid stabilised with IP6
Last updated date: Jun 8, 2022 Views: 722 Forks: 0
Derrick Lau1, Chantal L. Márquez1,2, Michael W. Parker3, Till Böcking1, *
1EMBL Australia Node in Single Molecule Science and ARC Centre of Excellence in Advanced Molecular Imaging, School of Medical Sciences, University of New South Wales, Sydney, Australia; 2Laboratory of Molecular and Cellular Virology, Institute of Biomedical Sciences, Faculty of Medicine, Universidad de Chile, Santiago, Chile; 3St. Vincent’s Institute of Medical Research, Australia; Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Australia.
*For correspondence: till.boecking@unsw.edu.au
1. Background
The human immunodeficiency virus (HIV) enters target cells by membrane fusion and releases the viral capsid containing HIV genomic RNA and associated proteins into the cytoplasm. The capsid is a conical protein container consisting of ~1500 copies of the viral capsid protein (CA) that assemble into a closed lattice of hexamers and pentamers. This lattice is stabilised by the cellular metabolite inositol hexakisphosphate that binds to a highly conserved ring of arginine residues in the centre of CA hexamers. The disassembly of the HIV-1 capsid, also known as uncoating, needs to occur at the right place and time for a productive infection to occur.
To study the properties of the HIV capsid and its interactions without the need for low-yielding capsid isolation procedures, we have recently developed a method to permeabilize HIV particles with a perfringolysin O (PFO), a cholesterol-dependent cytolysin that assembles into ring-shaped oligomers on cholesterol-containing membranes to form large transmembrane pores with a diameter of ~30–40 nm. These pores serve as ‘windows’ that allow passage of proteins in and out of the viral particle, facilitating real-time imaging of the interactions between the capsid and host proteins and of capsid uncoating by single-molecule fluorescence microscopy.1 The same approach has also been used to visualise the capsid by negative staining transmission electron microscopy (TEM) 1 and by cryogenic electron microscopy.2 Here, we describe the protocol for permeabilising HIV-1 particles with PFO followed by negative staining TEM. Our images reveal that PFO forms rings on the membrane of these particles containing IP6-stabilised capsids.
The HIV-1 particles used in this protocol lack the viral envelope protein and are non-infectious. Alternatively, virus-like particles prepared using lentiviral packaging plasmids can be used. Permeabilisation can be achieved using other pore-forming proteins in place of PFO, e.g. streptolysin O (SLO), which is commercially available.
2. Materials
A. HIV-1 particle preparation
- T25 cell culture flask
- Microwell plate
- 10 cm2 culture dishes (BD Biosciences, 353803)
- Poly-L-lysine-coated glass-bottom (175 μm thickness) 96-well plates (Greiner Sensoplate, Sigma, M4187)
- HEK-293T cells (ATCC, CRL-3216)
- Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Invitrogen, 11965-092)
- Fetal bovine serum (FBS) (Sigma-Aldrich, F2442-500ML)
- 1x PBS (Gibco, 10010031)
- 1x trypsin-EDTA (Gibco, 15400054)
- Plasmid, psPAX2 (NIH AIDS Reagent Program, 11348)
- Plasmid, pNL4.3-iGFP-∆Env (Hubner et al., 20073; Aggarwal et al., 2012)4
- Polyethylenimine (PEI Max) reagent (Polysciences, 9002-98-6)
- Sodium chloride solution 0.9% w/v (Sigma-Aldrich, S8776)
- HEPES (Sigma-Aldrich, H3375-250G)
- NaCl (Chem Supply, SA046-5KG)
- Cell culture media (DMEM supplemented with 10% fetal bovine serum)
- HEPES buffer saline pH 7.5 (HBS, 50 mM HEPES, pH 7.5, 100 mM NaCl)
B. Negative staining
- Carbon Type B electron microscope grid (Ted Pella, 01811)
- Corning Cell-tak cell tissue adhesive (Corning, 354240)
- Amicon concentrator Ultra-4, 3K MWCO (Millipore, UFC800324)
- 1 M NaOH
- 0.1 M sodium bicarbonate
- Uranyl formate 1% w/v
- Phytic acid sodium salt hydrate, also known as inositol-6-phosphate (IP6) (Sigma Aldrich, P8810-10G)
- Perfringolysin O (PFO, 20 µM in HBS)
- Whatman filter paper (Whatman, 1440-090) cut into pizza slices
- Bacterial petri dish
- Parafilm
- EM grid box
C. General
- Microcentrifuge tubes (Eppendorf, or equivalent)
- Milli-Q Water (from Millipore Milli-Q Integral 5 Water Purification System, or equivalent)
- 0.22 µm syringe filters
- Pipette tips (2, 200, 1000 μL)
3. Equipment
A. HIV-1 particle preparation
- 10-mL super loop (GE Healthcare, 18-1113-81)
- Tissue culture incubator, humidity, temperature and CO2 regulated (Thermo Fisher Scientific, model: 3110, or equivalent)
- Biosafety cabinet (Thermo Fisher Scientific, model: 1323TS, or equivalent)
- HiPrep 16/60 Sephacryl S-500 HR column (GE Healthcare, 28-9356-06)
- Fast protein liquid chromatography (FPLC) system including injector, one pump, UV-detector, fraction collector (GE Healthcare, ÄKTA pure, or equivalent)
- 4 °C refrigerator
- -20 °C freezer
- -80 °C freezer
B. Negative Electron microscopy staining and imaging
- Pelco easiGlow 91000 Discharge Cleaning System or equivalent plasma cleaner
- Pelco easiGlow TEM grid holder blocks (ProSciTech, PEL16820-81)
- TEM grid holder block (Pelco, PEL16820-81)
- Reverse action electron microscope grid tweezers (EMS Style 3X) or similar
- Water bath set at 37 °C with floaties
- FEI Tecnai G2 20 TEM operating at 200 kV
4. Procedures
A. Part I: HIV-1 viral particle preparation
Note: This protocol is reproduced from Márquez et al.5
- Produce HIV-1 viral particles (2.5 days)
Note: The combination of plasmids used in this protocol results in the production of viral particles that lack envelope proteins and are non-infectious; procedures should be done in accordance with local biosafety regulations for producing virus-like particles.- In a 15 mL conical tube, prepare DNA solution for transfection by mixing 6.6 μg of pNL4.3-iGFP-∆Env plasmid, 3.3 μg of psPAX2 plasmid, 60 μL of 1 mg/mL PEI max solution in a final volume of 500 μL of 0.9% (w/v) sodium chloride. Incubate the mixture for 30 min at room temperature to allow formation of DNA:PEI complexes.
- Split the HEK-293T cell culture as follows:
- Remove the culture medium and wash the cell monolayer with 1x PBS. Add 1x trypsin-EDTA (typically 1 mL for a T25 cell culture flask) and incubate for 5 min at 37 °C.
- Stop the trypsin digestion by adding new culture media and transfer the cell suspension to a 15 mL conical tube.
- Take a sample to count cells and determine the total number of cells in the tube. Centrifuge the cell suspension at 300 x g for 5 min at room temperature to pellet the cells and discard the supernatant. Resuspend the cells in the appropriate volume of fresh culture media to obtain a concentration of 7 x 106 cells/mL.
- Gently add 1 mL of cell suspension to the DNA:PEI mix and incubate for 5 min at room temperature.
- Plate the cells:DNA:PEI mixture drop by drop in a 10 cm2 culture dish containing 6.5 mL of culture media. Slightly shake the dish to distribute the cells homogeneously. Incubate at 37 °C with 5% CO2 for 48 h.
- Collect the virus-containing supernatant in a 15 mL conical tube and centrifuge at 2100 x g for 10 min at 4 °C to remove cellular debris.
- Collect the supernatant and transfer it to a new conical tube. The final volume of the cleared virus-containing medium should be around 7 mL.
Note: To verify the presence of fluorescent HIV particles in the supernatant, dilute 1 in 100 in PBS and then spinoculate 200 μL of the virus preparation onto a poly-L-lysine-coated glass-bottom 96-well plate well at 1,200 x g for 60 min at 4 °C, and inspect for fluorescent dots with a 60x objective in a fluorescence microscope. The number of viral particles per surface area can be determined by counting the number of fluorescent dots in at least 4 fields of view. The particle concentration can then be obtained by multiplying the number of particles per surface area with the surface area of the well (to obtain the number of particles per well) and then dividing this number by the volume of supernatant added to the well (0.2 mL).
- Purify the HIV-1 viral particles by size exclusion chromatography (1 day)
- Connect a HiPrep 16/60 Sephacryl S-500 HR size exclusion chromatography column to the FPLC system. All solutions used for FPLC should be passed through a filtration membrane with 0.2 μm pore size for removal of particulates and to degas the solutions.
- Replace the storage solution (usually 20% ethanol) with 2 column volumes (CV) of purified water and then equilibrate the column with 2 column volumes (CV) of HBS pH 7.5. Monitor the absorption at 280 nm. Flow rate: 1 mL/min.
Note: This step should be started the day before collecting the viral particles as it takes ~8 h for the column to be equilibrated. - Load the viral particles (~6.5 mL) onto the column using a 10-mL super loop. Monitor the absorption at 280 nm. Flow rate: 0.5 mL/min.
- Elute the sample with HBS pH 7.5 and collect 1 mL fractions until a total of 1.5 CV is reached (~6 h). A representative elution profile is shown in Figure 1.
- Combine the fractions corresponding to the first small peak that contain the viral particles (typically around fractions 34-39 or C10-D3 on a microwell plate, see Figure 1).
- Purified viral particles can be used within 7 days if they are stored at 4 °C, or you can make 200-500 μL aliquots and store at -80 °C (no flash freezing in liquid nitrogen needed). Frozen samples can be thawed on ice for use.
- Wash the column with 2 CV of purified water and then with storage solution.
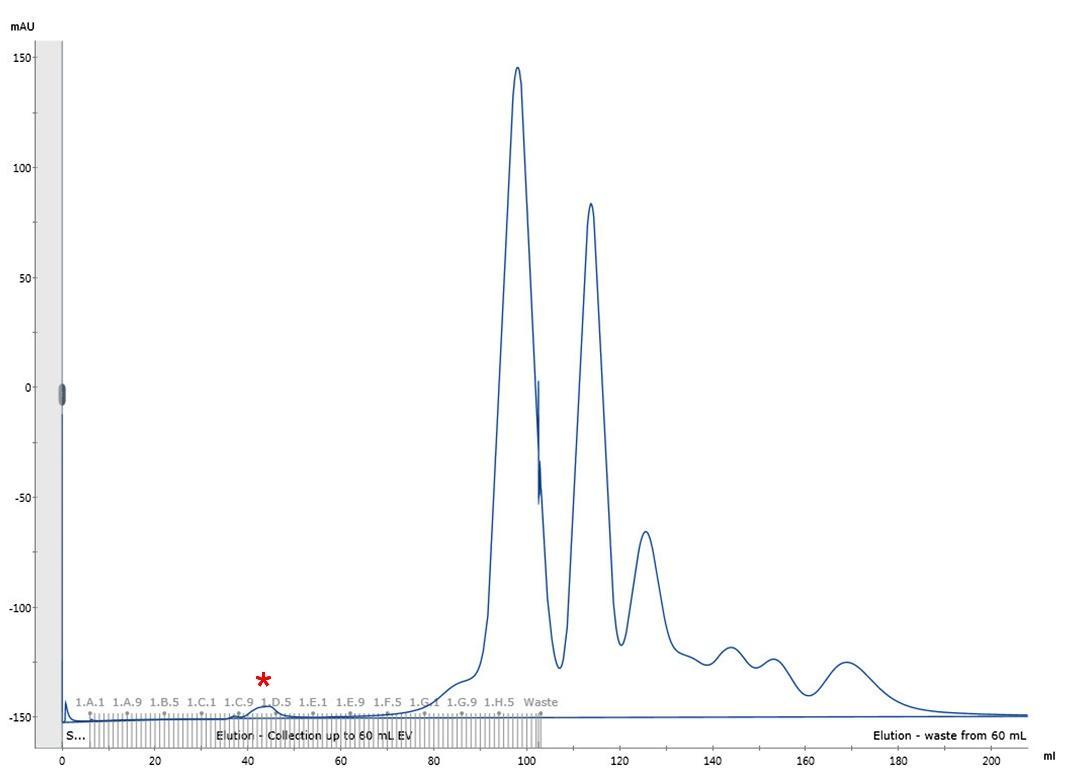
Figure 1. Representative elution profile of HIV-1 viral particles by size exclusion chromatography. Chromatographic separation of HIV-1 viral particles from culture media proteins. The first small peak (fractions C10-D3, marked with a red asterisk) corresponds to the HIV-1 viral particles. Reproduced from Márquez et al.5
B. Part II: Negative staining of viral particles stabilised with IP6
- Preparing reagents for negative staining
- Thaw viral particles and concentrate the particles 40–60-fold using the Amicon concentrator Ultra-4, 3K MWCO at 4000 x g, 4 °C at 10 min per spin. Resuspend after each cycle to prevent clumping until the final volume is achieved.
Note: For example, the pooled SEC fractions C10-D3 (~9.5 mL) in Figure 1 were concentrated to a final volume of 160 μL. - Thaw a solution of uranyl formate (1% w/v in water) and centrifuge at 18000 x g, 30 min at room temperature to remove precipitate. Transfer the supernatant to a clean Eppendorf tube wrapped in aluminium foil to protect uranyl formate from light.
- Prepare the Corning Cell-tak cell tissue adhesive solution (100 μL) by mixing 3.34 μL of 1.5 mg/mL of Cell-tak tissue adhesive solution (Corning, 354240) + 93 μL of 0.1 M sodium bicarbonate + 1.73 μL of 1 M NaOH.
- Dilute perfringolysin O in HBS to a final concentration of 20 µM and keep it at room temperature.
- Thaw viral particles and concentrate the particles 40–60-fold using the Amicon concentrator Ultra-4, 3K MWCO at 4000 x g, 4 °C at 10 min per spin. Resuspend after each cycle to prevent clumping until the final volume is achieved.
- Passivating the electron microscope grid with Cell-tak
- Pick up Carbon Type B electron microscope grid (Ted Pella, 01811) with reverse action tweezers and place them onto the Pelco easiGlow TEM grid holder block (Pelco, PEL16820-81) with the carbon (dark) side facing up.
- Insert the grid holder block into the Pelco easiGlow 91000 Discharge Cleaning System and glow discharge the grid on AUTO setting.
- Apply 5 μL of the cell-tak adhesive solution onto a glow discharged EM grid on the carbon (dark) side of the grid and invert the grid on a layer of parafilm so that the grid floats upside down.
- Incubate the cell-tak on the EM grid for 5 min.
- Negative staining of viral particles
- Prepare viral particle solution by mixing 9.5 μL of concentrated viral particles + 0.25 μL of IP6 (4 mM) + 0.25 μL of PFO (20 µM). The final concentration of PFO is 500 nM and IP6 is 1 mM. This is sufficient for preparation of 2 grids.
Note: IP6 is prone to precipitation in the presence of divalent cation. If needed the IP6 concentration can be lowered to 0.1 mM or can be substituted with mellitic acid (hexacarboxybenzene) to stabilise the capsids. - Incubate the viral particle solution in a water bath (37 °C) for 10 min for PFO to form pores on the membrane.
- Wick dry a Cell-tak coated EM grid with a small piece of filter paper.
- Apply 5 μL of the incubated viral particles solution to the side coated with Cell-take and incubate at room temperature for 1 min.
- Wick dry the sample using filter papers.
- Apply 5 μL of uranyl formate (1%, supernatant, free from aggregates), wick dry immediately with filter paper.
- Repeat step 5 twice. Wick dry with filter paper and allow the sample to air dry and store at room temperature in an EM grid box.
- Load sample onto the FEI G2 Tecnai and image at condenser aperture of 3 and objective aperture of 1 or 3 at x19500 magnification. See Figure 2 for representative images.
- Prepare viral particle solution by mixing 9.5 μL of concentrated viral particles + 0.25 μL of IP6 (4 mM) + 0.25 μL of PFO (20 µM). The final concentration of PFO is 500 nM and IP6 is 1 mM. This is sufficient for preparation of 2 grids.
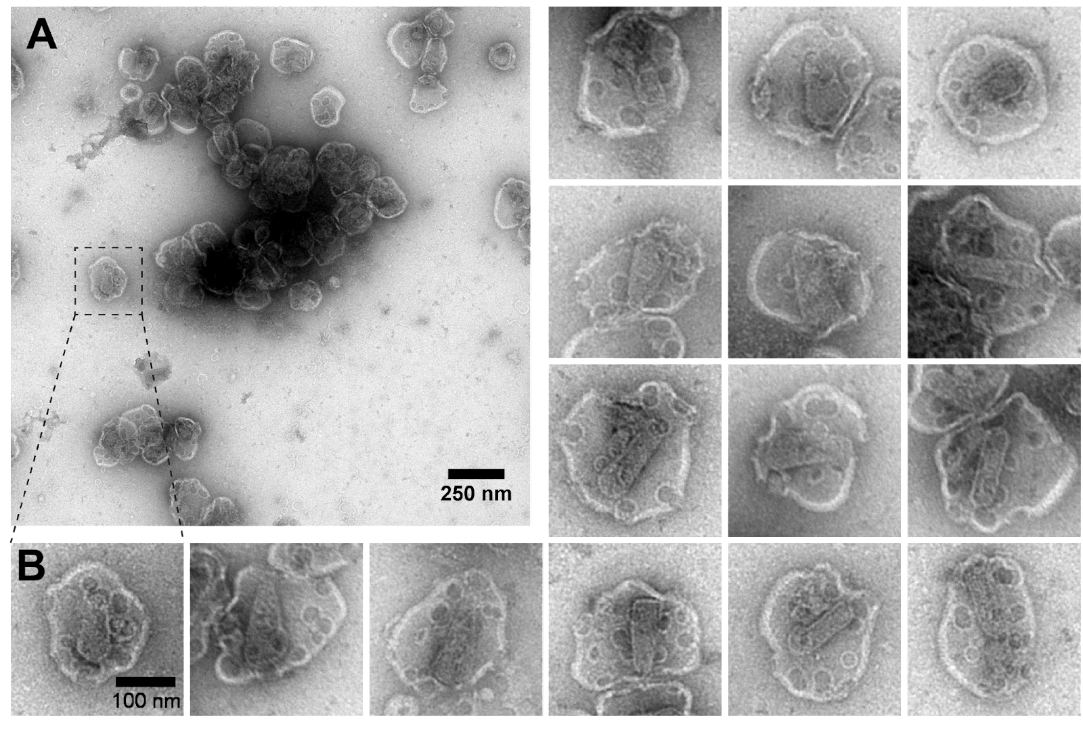
Figure 2. Representative electron micrographs of negative stained HIV particles treated with PFO and IP6. A. A typical field of view at 19500x magnification. Scale bar 250 nm. B. Selected HIV particles showing circular PFO pores and conical HIV-1 capsid inside. Scale bar 100 nm.
5. References
1. Márquez, C. L.; Lau, D.; Walsh, J.; Shah, V.; McGuinness, C.; Wong, A.; Aggarwal, A.; Parker, M. W.; Jacques, D. A.; Turville, S.; Böcking, T., Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. Elife 2018, 7, e34772.
2. Ni, T.; Zhu, Y.; Yang, Z.; Xu, C.; Chaban, Y.; Nesterova, T.; Ning, J.; Böcking, T.; Parker, M. W.; Monnie, C.; Ahn, J.; Perilla, J. R.; Zhang, P., Structure of native HIV-1 cores and their interactions with IP6 and CypA. Sci Adv 2021, 7 (47), eabj5715.
3. Hübner, W.; Chen, P.; Del Portillo, A.; Liu, Y.; Gordon, R. E.; Chen, B. K., Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol 2007, 81 (22), 12596-607.
4. Aggarwal, A.; Iemma, T. L.; Shih, I.; Newsome, T. P.; McAllery, S.; Cunningham, A. L.; Turville, S. G., Mobilization of HIV spread by diaphanous 2 dependent filopodia in infected dendritic cells. PLoS Pathog 2012, 8 (6), e1002762.
5. Márquez, C. L.; Lau, D.; Walsh, J.; Faysal, K. M. R.; Parker, M. W.; Turville, S. G.; Böcking, T., Fluorescence Microscopy Assay to Measure HIV-1 Capsid Uncoating Kinetics in vitro. Bio Protoc 2019, 9 (13), e3297.
- Lau, D, Márquez, C L, Parker, M and Böcking, T(2022). Negative staining of HIV viral particles permeabilised with PFO with capsid stabilised with IP6. Bio-protocol Preprint. bio-protocol.org/prep1709.
- Márquez, C. L., Lau, D., Walsh, J., Shah, V., McGuinness, C., Wong, A., Aggarwal, A., Parker, M. W., Jacques, D. A., Turville, S. and Böcking, T.(2018). Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. eLife. DOI: 10.7554/eLife.34772
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

