Advanced Search
ATAC-seq methodology used in Dong et al., eLIFE, 2020
Last updated date: May 3, 2022 Views: 717 Forks: 0
ATAC-seq detailed protocol by Xiaoyun Xing (Ting Wang Lab, WUSTL) As reported in Dong et al., eLIFE, 2020 (PMID: 32048992)
Prior to Transposition:
Make sure the cells are viable!
Recommend DNase I treatment for primary cells prior to nuclei isolation
Procedure
- Centrifuge the cells in a 1.5-ml microcentrifuge tube at 500 rcf for 10 min at 4 °C
- Remove supernatant without disrupting the pellet using two pipetting steps (aspirate down to 100 μl with a p1000 pipette and remove final 100 μl with a p200 pipette) and resuspend the pellet in 300 μl DNase Solution.
- Pipette mix 5x and incubate on ice for 5 min.
- Add 1 ml PBS + 0.04% BSA.
- Centrifuge cells at 500 rcf for 10 min at 4°C.
- Remove supernatant without disrupting the pellet and resuspend the pellet in 1 ml PBS + 0.04% BSA.
- Repeat steps d-e for a total of 2 washes.
- Pellet cells at 500 RCF at 4°C for 5 min in a fixed angle centrifuge.
- Aspirate all supernatant, carefully avoiding visible cell pellet, using two pipetting steps.
- Add 100 μl cold ATAC-Resuspension Buffer (RSB) containing 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin and pipette up and down to mix well.
- Incubate on ice for 3 minutes.
- Wash out lysis by adding 1 ml of cold ATAC-RSB containing 0.1% Tween-20 but NO NP40 or digitonin to the tube and invert tube 3 times to mix.
- Pass nuclei through a 30 μm Cell Strainer if there are nuclei clumps.
- Pellet nuclei at 500 RCF for 5 min at 4°C in a fixed angle centrifuge.
- Aspirate all supernatant using two pipetting steps.
- Resuspend cell pellet in 20 μl 2x TD buffer by pipetting up and down 6 times.
- Count nuclei using Trypan blue and a hemocytometer and transfer 50,000 nuclei into a new tube.
- Adjust nuclei to 25 ul total volume using 2x TD buffer.
- Add 25 μl of Omni-ATAC ATAC-seq reaction mix (2.5 μl Tagment DNA Enzyme 1, 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, 5 μl H2O) to the 25 μl of 50,000 nuclei. Pipet up and down 6 times.
- Incubate reaction at 37°C for 30 minutes in a thermomixer with 1000 RPM mixing. (Or in a heat block, mix by taping the tube every 10 min during the incubation)
- Cleanup reaction with a Zymo DNA Clean and Concentrator-5 Kit.
- Elute DNA in 21 μl elution buffer and store at -20°C until ready to amplify. This elution typically results in ~20 μl of product. Use all 20 μl of product in the following PCR.
- Amplify for 9 cycles using NEBNext 2x MasterMix:
9-12 cycle Amplification | |
| 10 uM Nextera Primer1 | 2.5 μl |
| 10 uM Nextera primer2 with index code | 2.5 μl |
| 2x NEBNext Master Mix | 25 μl |
| Transposed Sample | 20 μl |
Cycling Conditions | |
| 72°C | 5 min |
| 98°C | 30 sec |
Then 9 cycles of: | |
| 98°C | 10 sec |
| 63°C | 30 sec |
| 72°C | 1 min |
Hold at 4°C | |
24. Size selection with Ampure XP beads: follow the next steps.
25. Add 27.5 μl beads (0.55 x sample volume) and mix 10 times by pipetting.
26. Incubate 5 min at room temperature. Separate on magnetic stand.
27. Transfer supernatant (77.5 μl) to a new tube.
28. Add 50 μl beads (beads 27.5μl+50μl=77.5μl, 77.5μl/50μl initial sample=1.55 x sample volume).
29. Incubate 5 min at room temperature. Separate on magnetic stand.
30. Wash beads 2 times with 200 μl ethanol (80 %).
31. After the second wash, remove ethanolcompletely and put beads on a 37°C heat block with the caps opened until beads are dry, then resuspend beads in 12 μl EB by pipetting 10 times.
32. Separate on magnetic stand and transfer the final library to a new tube.
33. Quantify final libraries using Qubit (1 μl/sample, Qubit dsDNA HS Assay Kit) and check for library size distribution using 4200 TapeStation (High Sensitivity D1000 ScreenTape and Reagents).
34. Samples are now ready for sequencing on Illumina platforme.g., NextSeq (75 bpPE).
Buffer Preparations
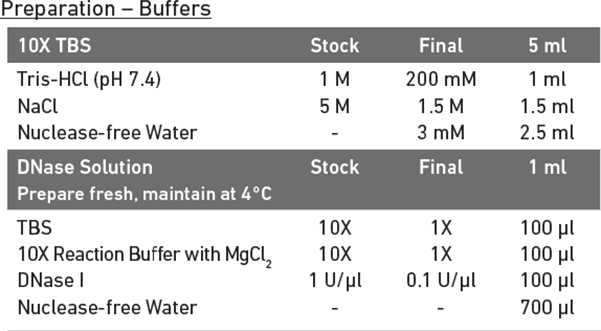
ATAC-RSB
| Reagent | Final Conc. | Vol for 50 ml |
| 1M Tris-HCl pH 7.4 | 10 mM | 500 μl |
| 5M NaCl | 10 mM | 100 μl |
| 1M MgCl2 | 3 mM | 150 μl |
| H2O | 49.25 ml |
Detergents - All detergents are resuspended as 100x stock solutions
- Digitonin: Digitonin is supplied at 2% in DMSO. Dilute 1:1 with water to make a 1% (100x) stock solution. Store at -20°C.
- Tween-20: Tween-20 is supplied at 10%. Use at this concentration (100x stock). Store at 4°C.
- NP40: NP40 is supplied at 10%. Use at this concentration (100x stock). Store at 4°C.
2x TD Buffer
| Reagent | Final Conc. | Vol for 100 ml |
| 1M Tris-HCl pH 7.6 | 20 mM | 2 ml |
| 1M MgCl2 | 10 mM | 1 ml |
| Dimethyl Formamide | 20% | 20 ml |
| H2O | 77 ml |
Aliquot and store in -20°C
Order List:
- DNase I, RNase-free includes10x Reaction Buffer with MgCl2 (ThermoFisher Scientific, EN0521)
- Trypan Blue (Thermo, 15250061)
- NP40 (Roche/Sigma, 11332473001)
- Tween-20 (Sigma/Roche, 11332465001)
- Tris pH 7.4 (Boston BioProducts, BBT-74)
- NaCl (Invitrogen/Thermo, AM9759)
- MgCl2 (Ambion/Thermo, AM9530G)
- Digitonin (Promega, G9441)
- PBS/DPBS (10010-023, 21040CM/21031CM)
- Tris pH 7.6 (Boston BioProducts, BBT-76)
- Dimethyl Formamide (Sigma, D4551-250ML)
- H2O (Corning, 46-000-CM)
- Tagment DNA Enzyme 1 (Illumina, FC-121-1030)
- 2x NEBNext Master Mix (NEB, M0541)
- DNA Clean and Concentrator-5 Kit (Zymo, D4014)
- Ampure XP beads (Beckman Coulter, A63880)
- Ethanol (Sigma, E7023-500ml)
- Qubit dsDNA HS Assay Kit (Thermo, Q32851)
- 4200 TapeStation High Sensitivity D1000 ScreenTape (Agilent, 5067-5584)
- 4200 TapeStation High Sensitivity D1000 Reagents (Agilent, 5067-5583)
Reference: Nature Methods: doi:10.1038/nmeth.4396. Corces et al. Nature Methods 2017
- Xing, X, Dong, C, Theunissen, T and Wang, T(2022). ATAC-seq methodology used in Dong et al., eLIFE, 2020. Bio-protocol Preprint. bio-protocol.org/prep1651.
- Dong, C., Beltcheva, M., Gontarz, P., Zhang, B., Popli, P., Fischer, L. A., Khan, S. A., Park, K., Yoon, E., Xing, X., Kommagani, R., Wang, T., Solnica-Krezel, L. and Theunissen, T. W.(2020). Derivation of trophoblast stem cells from naïve human pluripotent stem cells. eLife. DOI: 10.7554/eLife.52504
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
