Improve Research Reproducibility A Bio-protocol resource
Improve Research Reproducibility A Bio-protocol resource
Advanced Search
Preprint
LRP10 CRISPR/Cas9-mediated knockout in HuTu 80 cells
Last updated date: Mar 14, 2022 Views: 686 Forks: 0
LRP10 CRISPR/Cas9-mediated knockout in HuTu 80 cells (ATCC® HTB-40™)
Required materials
| Product | Brand and catalogue no. |
| DMEM/F-12 | Gibco™; 11320033 |
| Fetal Bovine Serum (FBS) | Gibco™ |
| Trypsin-EDTA (0.05%), phenol red | Gibco™; 25300054 |
| DPBS, no calcium, no magnesium (DPBS -/-) | Gibco™; 14190144 |
| pSpCas9-(BB)-2A-GFP (PX458) | Addgene; 48138 |
| BbsI | NEB; R0539S |
| NEBuffer™ r2.1 | NEB; B7030S |
| T4 DNA Ligase | NEB; M0202S |
| T4 DNA Ligase Reaction Buffer | NEB; B0202S |
| GeneJuice | Merck Millipore; 70967 |
| Hoechst 33342 solution | Invitrogen™; H3570 |
| HuTu-80 cells | ATCC® HTB-40™ |
| DMSO | Sigma-Aldrich; W387509 |
sgRNA in silico design for the LRP10 locus
- Find the CRISPR/Cas9 target sites using the CHOPCHOP web tool (http://chopchop.cbu.uib.no) or another preferable CRISPR/Cas9 design tool [1]. Pick single guide RNAs (sgRNA, 20 bp) with the highest predicted efficiency and the lowest predicted off-targets. We recommend testing the targeting efficiency of several sgRNA designed against different exons of the gene of interest.
- To clone sgRNA into pSpCas9-(BB)-2A-GFP [2], order sense and antisense oligonucleotides (Integrated DNA Technologies) with proper BbsI overhangs. These oligonucleotides need to be phosphorylated at the 5’ sites. Importantly, the pSpCas9-(BB)-2A-GFP allows the expression of the sgRNA by the human U6 promoter, which requires a “G” base at the transcription start site.
Add additional “G” at the start of the sgRNA sequence. For the LRP10 knockout, we used sgRNA targeting the first exon of the LRP10 gene (5′-P-CACCGCGTTTCGGTTCTTACCAAGG & 5′-P- AAACCCTTGGTAAGAACCGAAACGG).
sgRNA-oligo-FW: 5’-P-CACC(G)N1NNNNNNNNNNNNNNNNNNN20-3’ sgRNA-oligo-RV: 5’-P-AAACNNNNNNNNNNNNNNNNNNNN(C)-3’
Cloning of sgRNAs into pSpCas9-(BB)-2A-GFP plasmid
- Resuspend oligonucleotides to 100 µM with sterile demineralised water (dH2O) or 1X TE buffer and combine in the annealing reaction (see below). Incubate the reaction in the thermocycler at 95°C for 5 min. Then ramp down to 25°C at the speed of 5°C/min. The annealed product can be stored at -20°C.
1 µL of sgRNA-oligo-FW (final 10 µM) 1 µL of sgRNA-oligo-RV (final 10 µM)
1 µL of 10X T4 ligation buffer (final 1X) 7 µL of dH2O
Total volume: 10 µL - Set up the digestion for pSpCas9-(BB)-2A-GFP plasmid. Incubate at 37°C for 90 minutes. Inactivate the restriction enzyme for 20 min at 65°C. Load the digested vector on 1% agarose gel to check whether the restriction has worked. Typically, we notice high cutting efficiency of BbsI and therefore, we do not extract the linearised vector from the gel.
1 µL of 1 µg pSpCas9-(BB)-2A-GFP 2 µL of NEBuffer™ r2.1
1 µL of Bbs1 16 µL of dH2O
Total volume: 20 µL - Set up ligation. Before the ligation, dilute annealed oligonucleotides 1:250 in sterile dH2O. Incubate at 16°C overnight or at room temperature for 10 minutes. Heat inactivate at 65°C for 10 minutes. Chill on ice and transform 1-5 μL of the reaction into 50 μL competent cells according to your in-house bacterial transformation protocol. Plate the transformation on LB-Agar plates with 50 µg/mL ampicillin.
X µL of linearized pSpCas9-(BB)-2A-GFP (~50 ng) 2 µL of annealed oligonucleotides (1:250 dilution) 1 µL 10X T4 ligation buffer (final 1X)
0.5 µL T4 DNA ligase
Fill with dH2O to the total volume of 10 µL. - Next day, pick the colonies and inoculate in 3 mL LB medium containing 50 µg/mL ampicillin at 37°C and 200 RPM. Perform plasmid mini preparations and sequence to verify correct clones using a hU6-FW primer (5’- GAGGGCCTATTTCCCATGATT).
Cell culture and transfection of HuTu -80 cells
- Maintain HuTu 80 cells (ATCC® HTB-40™) in growth medium [DMEM/F-12 (Gibco), 10% (v/v) FBS, 1% Penicillin-Streptomycin] at 37°C/5% CO2. Split the cells when 80% confluent.
- Transfect the cells when 30-40% confluent in one well of a 6-well plate. Perform the transfection with pSpCas9-(BB)-2A-GFP containing sgRNA using GeneJuice transfection reagent according to the manufacturer’s specifications.
- 48 hours later, checkthe transfection efficiency using a fluorescent microscope. Transfected cells express GFP. Transfection efficiency in HuTu 80 cells is low to moderate (10-30%). Sort the cells 48 hours post-transfection.
Plate preparation
- Prepare 3X 96-well plates (flat bottom) by adding 150 µL of HuTu-80 growth medium. We normally culture the cells with Penicillin-Streptomycin after sorting, mainly to prevent contamination introduced during the cell sorting procedure.
- Warm up the plates before sorting at 37°C/5% CO2.
Clonal sorting of HuTu-80 cells
- For single-cell sorting, aspirate the medium from the well and wash once withDPBS-/-.
- Add 1 mL Trypsin-EDTA and place in the 37°C/5% CO2 incubator for 5 minutes.
- Add 1 mL growth medium to the well and transfer the cells to a 15 mL falcon tube.
- Spin the cells at 1000 RPM for 5 minutes.
- Aspirate the solution and resuspend the cells in 1 mL DPBS-/- containing 4% FBS.
- Transfer the cell solution to a round bottom tube with a cell strainer cap forsorting.
- Add 1 µL of sterile Hoechst 33342 solution and mix gently by inverting the tube.
- Sort GFP-positive cells into 3X 96-well plates (1 cell per well) through gating out doublets (forward and sideward scatter based) and dead cells (Hoechst 33342 positive cells). Figure 1 is an example of the Fluorescence-activated Cell Sorting (FACS) settings on BD FACSAria™ III for CRISPR/Cas9-mediated LRP10 KO in HuTu 80 cells.
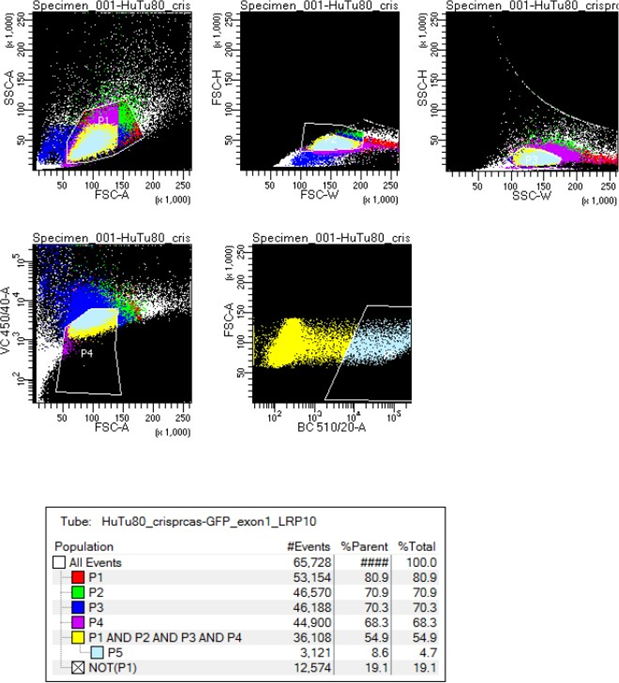
Figure 1 FACS sorting gating settings for CRISPR/Cas9-mediated LRP10 KO in HuTu 80 cells.
Expansion of single cells post sorting
- Right after sorting, incubate plates at 37°C/5% CO2 for 72 hours.
- Perform the first medium change with a 150 µL growth medium.
- Perform subsequent media changes every three days until colonies are observed.
- The colonies start to emerge 14 days after sorting. You can expect a minimum of 30 clones from three 96-well plates.
- When colonies reach 80% confluence, aspirate the media from the 96-well plates, wash with DPBS-/-, add 50 µL Trypsin-EDTA and incubate at 37°C/5% CO2.
- Next, add 150 µL growth media and transfer clones to separate 15 mL falcon tubes. Spin down at 1000 RPM.
- Resuspend the cells in fresh 150 µL growth media. Add one half of the cell suspension to a new well of a 48-well plate (PCR processing plate). Add another half of the cell suspension to a new 48-well plate (propagation plate).
- For the PCR processing plate: grow clones until they reach 80% confluence. When ready, extract DNA, perform PCR, Sanger sequencing, and INDEL tracking (TIDE, http://shinyapps.datacurators.nl/tide/) to identify targeted clones [3].
- For the propagation plate: grow clones until they reach 80% confluence and then cryopreserve as follows: aspirate the media, wash with DPBS-/-, add 50 µL of Trypsin-EDTA, and incubate at 37°C/5% CO2 for 5 minutes. To each well, add 300 µL growth media containing 10% DMSO. Wrap the plates with Parafilm™ wrapper and store in a zippered plastic bag n -80°C. The cells will stay frozen during the screening. According to your in-house protocol, as you identify positive clones, thaw and expand the desired clones.
References
- Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., & Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic acids research, 47(W1), W171-W174.
- Ran, F. A. F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., & Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nature protocols, 8(11), 2281-2308.
- Brinkman, E. K., Chen, T., Amendola, M., & van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic acids research, 42(22), e168-e168.
Copyright: Content may be subjected to copyright.
How to cite:
Readers should cite both the Bio-protocol preprint and the original research article where this protocol was used:
- Grochowska, M and Mandemakers, W(2022). LRP10 CRISPR/Cas9-mediated knockout in HuTu 80 cells. Bio-protocol Preprint. bio-protocol.org/prep1584.
- Grochowska, M. M., Mascaro, A. C., Boumeester, V., Natale, D., Breedveld, G. J., Geut, H., Cappellen, W. A. V., Boon, A. J. W., Kievit, A. J. A., Sammler, E., Parchi, P., Cortelli, P., Alessi, D. R., Berg, W. D. J. V. D., Bonifati, V. and Mandemakers, W.(2021). LRP10 interacts with SORL1 in the intracellular vesicle trafficking pathway in non-neuronal brain cells and localises to Lewy bodies in Parkinson’s disease and dementia with Lewy bodies. Acta Neuropathol 142(1). DOI: 10.1007/s00401-021-02313-3
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
0 Q&A
Spinning
This protocol preprint was submitted via the "Request
a Protocol" track.
Share
Bluesky
X
Copy link
Cancel
