Advanced Search
Study design
Last updated date: Dec 9, 2021 Views: 888 Forks: 0
we have uploaded the Appendix 1 containing the detailed protocol. In case you have further queries please drop a mail to debojyoti.chakraborty@igib.in
Appendix 1
RAY: a detection assay for major VOCs of SARS-CoV2
Materials and reagents
- Micropipettes
- Plastic ware (including sterile filter tips, PCR and micro centrifuge tubes).
- Personal protective equipment (PPE)
- General laboratory materials and reagents, commonly used in molecular biology labs
Equipment
- Microfuge / Centrifuge as necessary
- Thermal cycler
- Thermal bath (Heat block)
General precautions
- Set up the reactions in designated areas for sample processing, RT-PCR and amplification.
- Handle all RT-PCR amplified products in separate post-amplification areas to prevent contamination from amplified products.
- Use filter tips to set up all reactions.
- Set up positive control reactions at the end.
NOTE! | It is highly recommended to open tubes containing amplified PCR products from patient samples and positive controls, in a designated post amplification area, physically separated from the room where nucleic acid (RNA) extraction happens. |
Preparation of CRISPR-Cas9 crRNAs (to be performed prior to sample handling under RNase free condition):
1. Synthesis of in vitro transcribed (IVT) crRNAs targeting SAR-CoV2 S gene and N501Y mutation
- To assemble equimolar ratio of Forward and Reverse oligos (refer Table 1) for each target:
Reagents | Volume (µl) | Final Concentration |
Forward Oligo (100 µM) | 1.25 | 2.5 µM |
Reverse Oligo (100 µM) | 1.25 | 2.5 µM |
Total Volume (with nuclease free water) | upto 50 µl |
|
- Heat the reaction mix at 95oC for 5 min followed by slow cooling at room temperature for 15 min.
- Perform in vitro transcription using commercially available T7 Polymerase based IVT kit as per recommended protocol. A sample is given below (MEGAscript T7 Kit, ThermoFisher Scientific)
Assemble the following reaction components at room temperature:
Reagents | Volume (µl) | Final Concentration |
Nuclease free water | upto 20 |
|
ATP (75 mM) | 2 | 7.5 mM |
GTP (75 mM) | 2 | 7.5 mM |
CTP (75 mM) | 2 | 7.5 mM |
UTP (75 mM) | 2 | 7.5 mM |
10X Reaction Buffer | 2 | 1X |
Enzyme Mix | 2 | - |
Annealed oligo duplex from step 1 | 5 | - |
- Incubate the reaction mix overnight at 37oC.
- Add 1 µl of Turbo DNAse in the reaction mix and incubate at 37oC for 30 min.
- Heat inactivate at 70oC for 10 min.
Optional: RNA can be visualized on a 2% agarose gel to check for its integrity.
- Column based RNA clean-up as per the commercially provided protocols (such as NucAway Spin Columns, AM10070, ThermoFisher Scientific)
2. Generation of chimeric gRNAs (crRNA:tracrRNA-FAM)
Reagents | Volume (µl) | Final Concentration |
IVT synthesized crRNA | - | 1 µM |
FAM labelled tracrRNA | - | 1 µM |
Annealing Buffer (100 mM NaCl, 50 mM Tris-Cl pH 8.0, 1 mM MgCl2) | upto 50 |
|
- Heat the reaction mix at 95oC for 5 min followed by slow cooling at room temperature for 15 min.
NOTE! | The crRNA and chimeric gRNA products can be produced in bulk and stored at -20oC for long term use. |
FELUDA based detection for COVID-19 (RT-PCR):
1. Extract patient RNA according to CDC recommendations.
2. Set-up single step RT and PCR reaction.
Assemble the reaction components as below (using Biotin labelled primers):
Reagents | Volume (µl) | Final Concentration |
Forward Biotinylated Primer (10 µM) | 0.2 | 200 nM |
Reverse Biotinylated Primer (10 µM) | 0.2 | 200 nM |
dNTPs (10 mM) | 0.1 | 100 µM |
10X Reaction Buffer (200mM Tris-Cl pH 8.4, 500mM KCl) | 1 | (1X) |
MgCl2 (50 mM) | 0.3 | 1.5 mM |
DMSO (100%) | 0.3 | 3% |
Taq DNA polymerase (5 U/µl) | 0.05 | 0.025 U/µl |
Reverse Transcriptase (200 U/µl) | 0.25 | 5 U/µl |
RNA sample (≥ 5 ng)* | As per sample |
|
RNase inhibitor (Optional, in case not present with RT enzyme) 20U/µl | 0.2 | 0.4 U/µl |
Total Volume (with nuclease free water) | upto 10 |
|
NOTE! | For a single set of reactions one non-template control (NTC) and one positive control (PC) should be included, to check for non-specific amplification. |
Optional: In order to confirm amplification after single step RT and PCR, a volume of 1-2µl from NTC and PC can be checked on a 2% agarose gel.
Reaction Conditions:
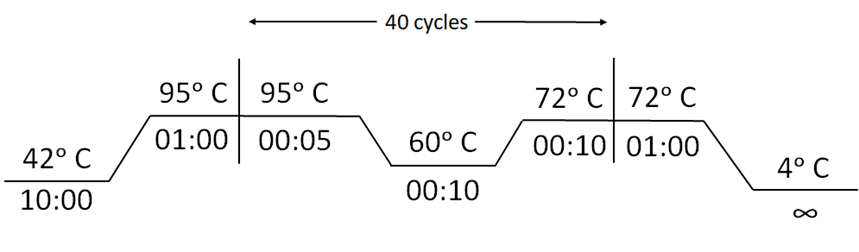
Appendix 1-figure 1: Cycling conditions for Reverse Transcription-PCR for RAY
3. Prepare dFnCas9-chimeric gRNA-RNP complexes for the samples to be tested. RNP complex against SARS-CoV-2 S gene and N501Y mutation should be assembled for each sample.
- Incubate dFnCas9 protein with Chimeric FAM labelled guide RNA to generate RNP complexes for 10 min at Room Temperature (RT).
Reagents | Volume (µl) | Final Concentration |
dFnCas9 protein (1µM) | 1.0 | 100 nM |
Chimeric FAM labelled gRNA (1 µM) | 1.0 | 100 nM |
Total Volume (with Buffer containing 20 mM HEPES pH 7.5, 150 mM KCl, 10% glycerol, 1mM DTT and 10mM MgCl2) | upto 5 |
|
4. Add 5 µL of the biotinylated amplicon (from Step 2) to 5µL of the dFnCas9 RNP complex (from Step 3)
NOTE! | For a single set of reactions one non-template control (NTC) and one positive control (PC) should be included, to check dFnCas9 RNP specificity. |
5. Incubate the reaction mix (containing RNP complex and amplicon substrate) at 37°C for 10 minutes in a heating block or water bath.
6. Add 80µL of dipstick buffer (provided) to each tube containing 10µL of the reaction mix from the previous step.
7. Insert Milenia HybriDetect 1 lateral flow strip directly into reaction tubes.
8. Allow the solution to migrate into the strip for 2 minutes at room temperature and observe the result.
- NTC (non-template control) to be used as negative control.
- For accurate results, positive samples show up in the test band within 2 min while negative samples show very weak or no signal.
- Incubating for longer times leads to increasing background signal intensity at test band location making interpretation ambiguous.
- Signals between positive and negative assays can also be interpreted by densitometry analysis from images captured using any photographic device.
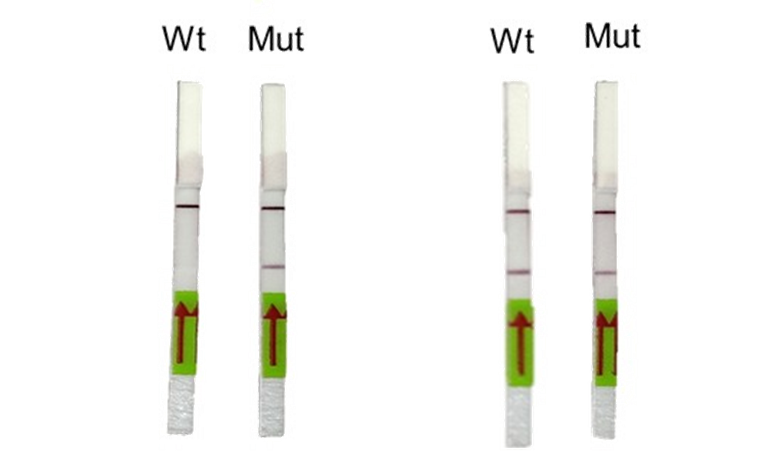
Appendix 1-figure 2: Representative image of RAY outcomes on a paper strip for sample having the N501Y mutation (Mut) or the parent CoV2 sequence (Wt). Left panel shows outcome with SN501Y sgRNA and the right panel shows the outcome with SWT sgRNA.
Image analysis through TOPSE smartphone app:
1. T.O.P.S.E (True Outcome Predicted via Strip Evaluation), starts by clicking “GET STARTED” on the home page, which further directs to an app page where access to previous recorded data is given through entering a operator ID (eg. XYZ) or a new session of data recording begins. The app then further asks to record a background value for No template control (NTC).
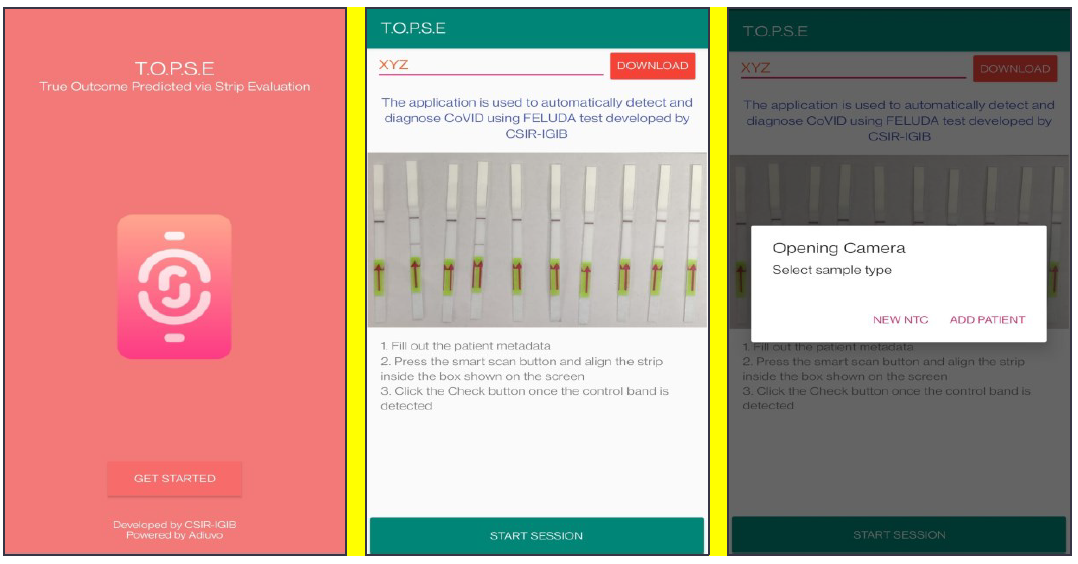
Appendix 1-figure 3: Representative screen-shots showing TOPSE app home page and user interface.
2. Upon selecting “NEW NTC”, a new app page having a strip holder pops up, once the strip is placed in the holder, a numerical band intensity for the test band is calculated to set a background value. After clicking on “DONE”, a pop up asks to record for a patient sample.
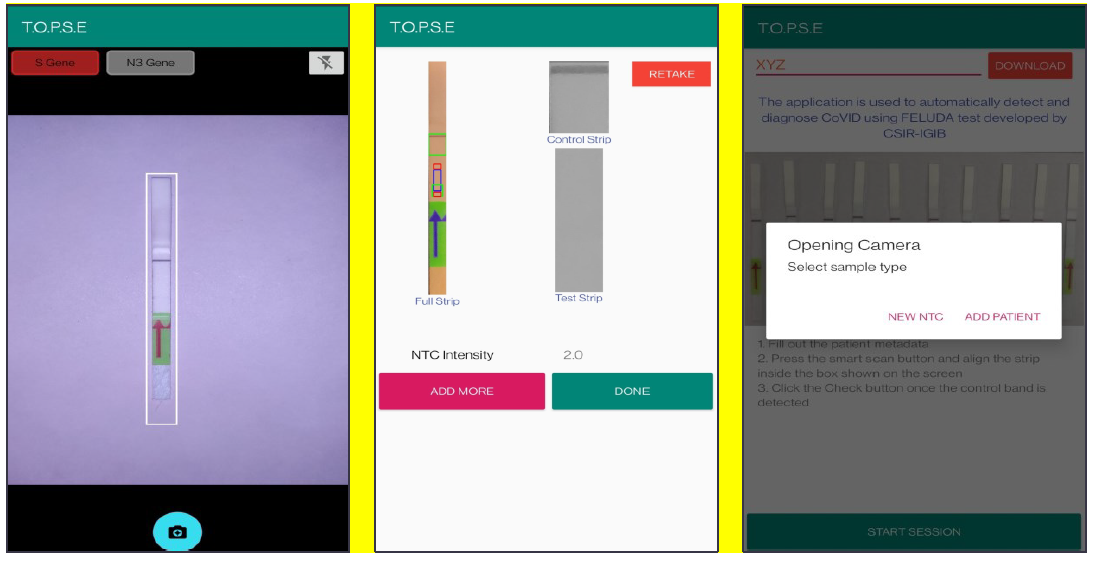
Appendix 1-figure 4: Representative screen-shots showing TOPSE image acquisition for NTC background correction.
3. In the next page, clicking on “OPEN CAMERA”, takes the operator to another strip holder to record test band intensity of the patient sample strip. If the calculated test band intensity is above NTC threshold, the sample will be called as “POSITIVE”.
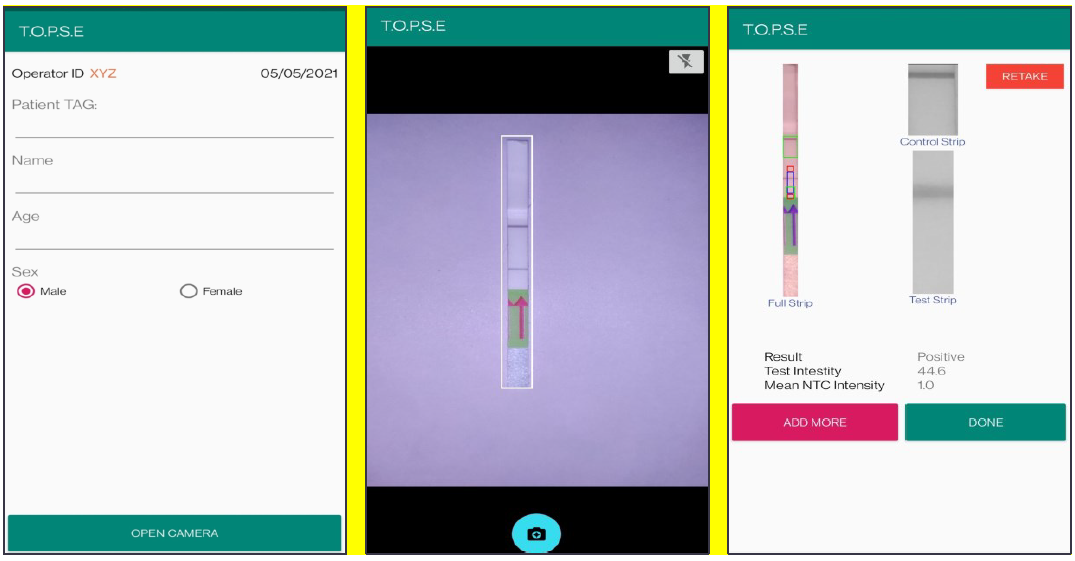
Appendix 1-figure 5: Representative screen-shots showing TOPSE data entry step and image acquisition for a sample identified as "Positive".
4. Once a recording session is done, one can record for another sample just by clicking onto “ADD PATIENT”, the app will automatically use the previous NTC threshold value and need to be set again. For eg. if the calculated test band intensity for the second sample is below NTC threshold or within the error limit of the app, the sample will be called as “NEGATIVE”.
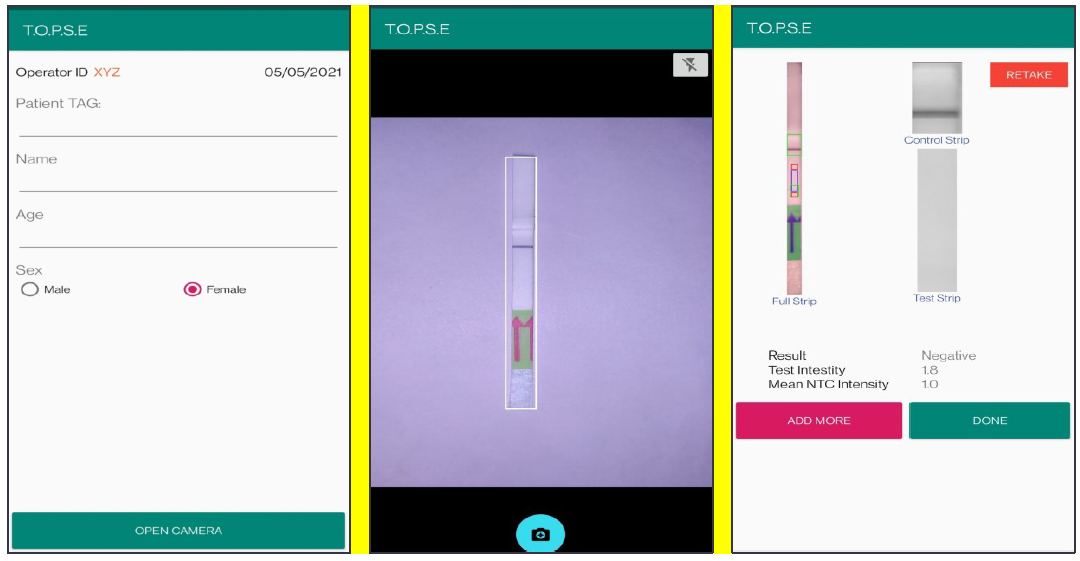
Appendix 1-figure 6: Representative screen-shots showing TOPSE data entry step and image acquisition for a sample identified as "Negative".
8. For using TOPSE to perform N501Y RAY assay, the test intensity of the sample needs to be >10.2 for a sample to be called positive for the N501Y assay. The S gene intensity has to be >6.7 for the sample to be called positive for SARS CoV2.
9. Strips may be discarded according to standard procedures.
List of primers and oligos are mentioned in Supplementary File 4.
Reagents used:
MEGAscript T7 Transcription Kit (Cat. No. AM1334, ThermoFisher Scientific), Quantitect RT (Cat. No. 205311, Qiagen), Taq DNA Polymerase (Cat. No. 18038018, ThermoFisher Scientific), 5’ end biotinylated primers (Merck, Darmstadt, Germany), 3' end 6 – Fluorescein amidite (6-FAM) FnCas9 tracrRNA (GenScript Biotech (New Jersey, USA) or Merck (Darmstadt, Germany), MILENIA HybriDetect1 (Cat. No. MILLENIA 01, TwistDx, UK or Giessen, Germany).
- Kumar, M, Maiti, S and Chakraborty, D(2021). Study design. Bio-protocol Preprint. bio-protocol.org/prep1465.
- Kumar, M., Gulati, S., Ansari, A. H., Phutela, R., Acharya, S., Azhar, M., Murthy, J., Kathpalia, P., Kanakan, A., Maurya, R., Vasudevan, J. S., S, A., Pandey, R., Maiti, S. and Chakraborty, D.(2021). FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. eLife. DOI: 10.7554/eLife.67130
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

