Advanced Search
Production and titration of SARS-CoV-2 spike pseudotyped virus using lentiviral system
Last updated date: Jul 18, 2021 Views: 922 Forks: 0
Production and titration of SARS-CoV-2 spike pseudotyped virus using lentiviral system
Abstract
Lentiviral vectors can be pseudotyped with various heterologous viral glycoproteins that modulate cellular tropism and cell-entry properties. The protocol outlined represents a safe, rapid and reliable method for production high-titer viral pseudotype particles with SARS-CoV-2 spike protein. D614G substitution and deletion of cytoplasmic tail 19 amino acids on spike protein (termed S-C19del) generated pseudoviruses with higher titer. This assay allows the study of the role of SARS-CoV-2 spike protein, development of receptor binding and entry assays, detection of neutralizing antibodies, and screening of viral entry inhibitors. Further, this pseudovirus assay can be readily adapted to the newly emerged SARS-CoV-2 variants.
Keywords: SARS-CoV-2, Spike, Pseudovirus, Titration, Neutralization assay
Background
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Due to the highly pathogenic nature of SARS-CoV-2, infectious SARS-CoV-2 must be handled in a biosafety level 3 (BSL-3) facility. Pseudotyped viruses are valuable tools for studying virulent or lethal viral pathogens. Pseudovirus-based assays have been widely used for the study of cellular tropism, receptor recognition, and viral inhibitors as well as evaluation of neutralizing antibodies (Nie et al. 2020). The most efficient and common viral vectors for the production of pseudotype viral particles are lentiviruses. Human immunodeficiency virus type 1 (HIV-1) is the most thoroughly studied of lentiviruses.
Here, we generated a luciferase (Luc)-expressing SARS-CoV-2 pseudovirus in the envelope-defective HIV-1 backbone. Lentiviral particles bud at the cytoplasmic membrane of host cells (Milone et al. 2018), whereas coronaviruses assemble and bud at membranes of the intermediate compartment between the endoplasmic reticulum (ER) and Golgi complex (Nguyen et al. 1997). Therefore, in the lentivirus-based pseudotype system, heterologous envelope glycoproteins need to be transported to the plasma membrane for incorporation into pseudotyped progeny virions. Deletion of 19 amino acids on the cytoplasmic tail domain promoted coronavirus spike protein transport to cytoplasmic membrane, which significantly improved the packaging efficiency (Ou et al. 2020).
This pseudovirus contains a Luc reporter gene that enabled us to easily quantify virus entry into human angiotensin-converting enzyme 2 (hACE2)-expressing HKE293T cells. This single-round pseudovirus assay is extremely safe and useful alternative to live virus-based assay without the need for high containment facilities. Figure 1 shows a cartoon of the lentiviral SARS-CoV-2 pseudovirus production process directed by plasmid co-transfection.
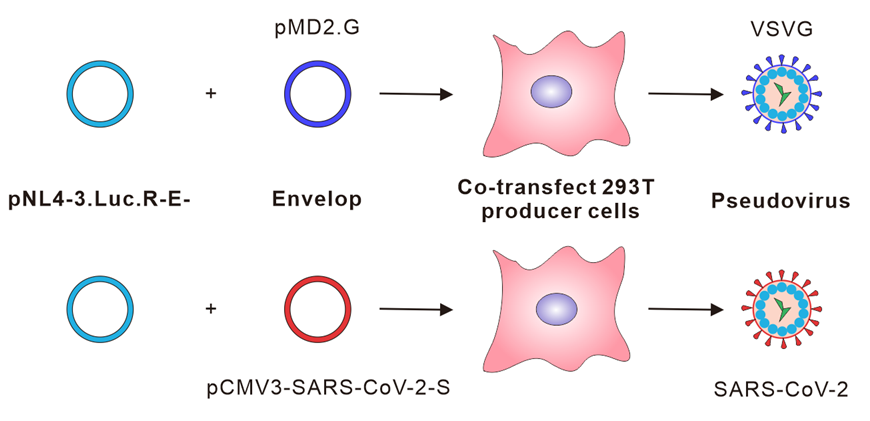
Figure 1. Schematic representation of the SARS-CoV-2 pseudovirus production. HEK293T cells are transfected with two plasmids, pNL4-3.Luc.R-E- (Connor et al. 1995) and SARS-CoV-2 spike expression plasmid, for the production of SARS-CoV-2 spike bearing pseudovirus.
Materials and Reagents
- Pipette tips (filtered) (AXYGEN, catalog numbers: TF-300-R-S, TF-200-R-S, TF-1000-R-S)
- 0.2 ml PCR tubes (AXYGEN, catalog number: PCR-02-C)
- 1.5 ml Microtubes (AXYGEN, catalog number: MCT-150-L-C)
- 50 ml tubes (Falcon, catalog number: 352098)
- 96-well plates (Corning, catalog number: 3599)
- 24-well plates (Corning, catalog number: 3524-ND)
- 6-well plates (Corning, catalog number: 3516)
- 10 cm dishes (Corning, catalog number: 430167)
- M02-ACE2 (Genecopoeia, catalog number: EX-U1285-M02-B)
- 0.45 μm filter (Millipore, catalog number: HVHP04700)
- TRIzol (Invitrogen, catalog number: 15596026)
- PrimeScript RT reagent Kit with gDNA Eraser (Takara, catalog number: RR047A)
- DMEM (Gibco, catalog number: C11995500CP)
- FBS (Gibco, catalog number: F8687-500ML)
- Penicillin-Streptomycin (HyClone, catalog number: SV30010)
- Opti-MEM (Gibco, catalog number: 31985062)
- PBS (HyClone, catalog number: SH30256.01)
- 0.25% Trypsin-EDTA (Gibco, catalog number: 25200072)
- Lipofectamine™ 3000 Transfection Reagent (Thermo, catalog number: L3000015)
- EndoFree Plasmid Mini Kit (CWBIO, catalog number: CW21065)
- SARS-CoV-2 spike plasmid (Sino Biological, catalog number: VG40589-UT)
- PrimeSTAR Max DNA Polymerase (Takara, catalog number: R045A)
- iQ SYBR Green Supermix (Bio-Rad, catalog number: 1725121)
- Xba I (Takara, catalog number: 1634)
- Kpn I (Takara, catalog number: 1618)
- T4 DNA Ligase (Takara, catalog number: 2011A)
- Luciferase Assay System (Promega, catalog number: E1500) (containing: Luciferase Assay Substrate E151A; Luciferase Assay Buffer E152A; Cell Culture Lysis 5× Reagent E153A)
Equipment
- PCR Authorized Thermal Cycler (Eppendorf AG, 22331 Hamburg)
- Eppendorf mixer (Eppendorf, catalog number: 5382000074)
- -80°C freezer (Thermo, catalog number: 905)
- CFX Connect Real-Time System (Bio-Rad, CFXTM Optics Module)
- Micro-Spectrometer (ALLSHENG, Nano-500)
- INCUBATOR SHAKER (CRYSTAL, IS-RSV1)
- Centrifuge (Eppendorf, model: 5810R & 5418R)
- GLOMAX MULTI JR DITECTION SYSTEM (Promega, Model: E6080)
Procedure
A. Lentiviral Production
Briefly, to make lentivirus, a transfer plasmid must be co-transfected into HEK293T cells with the packaging plasmids. It is essential that the cells be well-maintained and of relatively low passage number.
- One day before transfection, seed HEK293T cells in the 10cm plate. Maintain cells at 37°C and 5% CO2 in a humidified incubator.
(CRITICAL STEP: Cells should preferably be of low passage number) - The cells should be around 80% confluent at the time of transfection.
- Transfect 6μg SARS-CoV-2 spike plasmid and 6μg pNL4-3.Luc.R-E- into HEK293T cells using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s protocol. Meanwhile, transfect 2μg envelop plasmid VSV-G and 10μg pNL4-3.Luc.R-E- into HEK293T cells as control.
- After 8 h, replace the transfection medium with 10 ml of fresh DMEM + 10% FBS (Sigma). Incubate the cells at 37°C for 48~72 h. Importantly, transfection efficiency should be around 90%.
- Harvest the lentiviral supernatants into a 15 ml centrifuge tube and centrifuge briefly (2000 rpm, 8 min at 4°C), and filter through a 0.45-μm filter to remove cellular debris.
- Aliquot the virus (0.5~1 ml) and store at -80°C. Each freeze–thaw cycle may reduce the viral titers by up to two- to four-fold.
B. Lentiviral Particles Titration
(a) RNA extraction:
1. Take 400μl pseudotyped viruses VSVG, SARS-CoV-2 and D614G in separated RNase-free 1.5 ml tube, add 400μl TRIzol, vortex for 30 seconds.
2. Add 160μl chloroform, vortex for 30 seconds.
3. Centrifuge at 4 °C, 12000g for 10 min.
4. Transfer the supernatant to a new 1.5ml eppendorf tube.
5. Add equal volume of isopropanol, vortex and mix well.
6. Centrifuge at 4 °C, 12000g for 10 min.
7. Discard the supernatant and leave the precipitate.
8. Wash the precipitate with 75% ethanol and centrifuge at 4 °C, 12000g for 10 min, twice.
9. Use 7 μl of RNase-free water to dissolve the pellet.
(b) Reverse transcription
1. Add 2 μl 5×gDNA Eraser Buffer, 1μl gDNA Eraser to the 7 μl dissolved RNA.
2. Incubate the mixture at 42°C for 2 min.
3. Then ice bath for over 2 min.
4. Add PrimeScript RT Enzvme Mix l (1 μl), RT Primer Mix (1 μl), 5× PrimeScript Buffer 2 (for Real Time) (4 μl), RNase Free H2O (4 μl), total 20 μl.
5. Incubate the mixture at 37°C for 15 min, 85°C for 5 seconds, and then ice bath.
(c) Real-time qPCR
1. The titers of the pseudoviruses were calculated by determining the number of viral RNA genomes per mL of viral stock solution using qPCR with primers that target LTR (Geraerts et al. 2006). Sense primer: 5′-TGTGTGCCCGTCTGTTGTGT-3′, anti-sense primer: 5′-GAGTCCTGCGT CGAGAGAGC-3′.
2. 10 μl reaction system:5 μl iQ SYBR Green Supermix, 0.5 μl Sense primer, 0.5 μl anti-sense primer, 2 μl RNase Free H2O, 2 μl cDNA.
3. Dilute the pNL4-3.Luc.R-E- vector (5×108 to 5×102) copies/μl as standard.
4. Amplification conditions: 98°C for 5 min, (98°C for 15 seconds, 58°C for 10 seconds, 72°C for 15 seconds, read the plate, 40 cycles), Melt curve (55°C to 95°C).
5. Analyze the results with Graphpad 8.
6. Adjust the pseudoviruses to the same titer (copies/mL) for the following experiments.
C. Pseudovirus based neutralization assay
- Plate target cells (293T-hACE2) in a 96-well plate 12 hours prior to viral infection, 2×104 per well.
- The cells should be approximately 70% confluent on the day of infection. (Typically cells are transduced at 50~80% confluency.)
- Dilute the convalescent sera (1:20, 1:40, 1:80, 1:160, 1:320, 1:640, 1:1080, 1:2560, 1:5120) serial dilution in an empty, sterile 96-well plate.
- Add 50 μl pseudoviruses supplemented with polybrene (5 mg/mL) per well, and incubate the mixture for 1 h at 37°C, after incubation, remove cell maintenance media from the 293T-hACE2 96-well plate, then transfer to the mixture into the 96-well plate which containing target cells. Incubate the infected 293T-hACE2 cells for 12 h at 37°C in the 5% CO2 incubator.
- At 12 h post-infection (p.i.), remove infectious media and add fresh culture for another 60 h.
- At 72 h p.i., remove media and lyse the cells with Cell Culture Lysis for 10 min at 37°C, and measure the activity of firefly luciferase in cell lysates using a commercial substrate (Luciferase Assay System).
D. SARS-CoV-2 spike drive entry into human cell lines.
1. The treatment of HEK293T and 293T-hACE2.
2. Plate target cells (HEK293, 293T-hACE2) in a 96-well plate 12 hours prior to viral infection, 2×104 per well.
3. Add 50 μl pseudoviruses (VSVG, SARS-CoV-2 WT, D614G) supplemented with polybrene (5 mg/mL) per well, Incubate the infected 293T-hACE2 cells for 12 h at 37°C in the 5% CO2 incubator.
4. At 12 h post-infection (p.i.), remove infectious media and add fresh culture for another 60 h.
5. At 24h, 48h and 72h p.i., remove media and lyse the cells with 100 μl Cell Culture Lysis for 10 min at 37°C, and measure the activity of firefly luciferase in cell lysates using a commercial substrate (Luciferase Assay System). (80 μl cell lysis plus 80 μl Luciferase substrate).
Figure 2. The treatment of HEK293T and 293T-hACE2. We plate target cells (HEK293T, 293T-hACE2), add pseudoviruses (VSVG, SARS-CoV-2 WT, D614G), measure the activity of firefly luciferase in different time (24h, 48h and 72h).
6. The results are in the Figure 3.
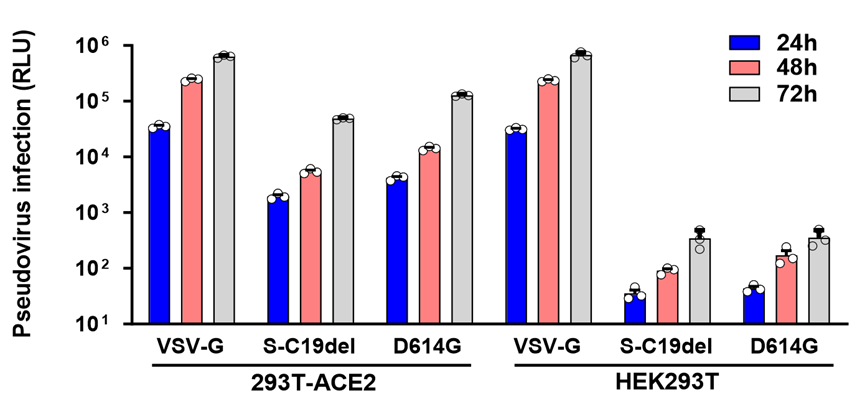
Figure 3. The entry efficiency was determined by quantifying luciferase activities of cells lysates. We plate target cells (HEK293T, 293T-hACE2), add pseudoviruses (VSVG, SARS-CoV-2 WT, D614G), measure the activity of firefly luciferase in different time (24h, 48h and 72h). The RLU showed that the infection of VSVG stay the same with or without the expressing of hACE2 in target cells. While SARS-CoV-2 and D614G cannot infect the HEK293T cells without the expressing of hACE2.
E. Pseudovirus based neutralization assay of convalescent sera
(1) Plate target cells (293T-hACE2) in a 96-well plate 12 hours prior to viral infection, 2×104 per well.
(2) Dilute the convalescent sera (1:40, 1:80, 1:160, 1:320, 1:640, 1:1080, 1:2560, 1:5120) serial dilution in an empty, sterile 96-well plate.
(3) Add 50 μl pseudoviruses supplemented with polybrene (5 mg/mL) per well, and incubate the mixture for 1 h at 37°C, after incubation, remove cell maintenance media from the 293T-hACE2 96-well plate, then transfer to the mixture into the 96-well plate which containing target cells. Incubate the infected 293T-hACE2 cells for 12 h at 37°C in the 5% CO2 incubator.
(4) At 12 h post-infection (p.i.), remove infectious media and add fresh culture for another 60 h.
(5) At 72 h p.i., remove media and lyse the cells with 100 μl Cell Culture Lysis for 10 min at 37°C, and measure the activity of firefly luciferase in cell lysates using a commercial substrate (Luciferase Assay System) (80 μl cell lysis plus 80 μl Luciferase substrate).
(6) The neutralization titters calculated as 50% inhibitory dose (ID50) are shown in Figure 4.
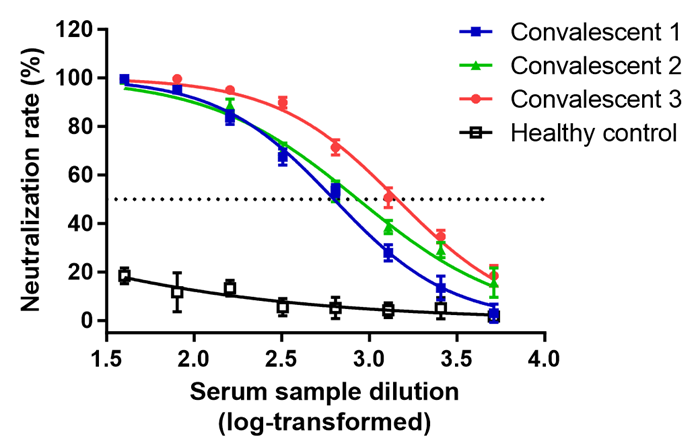
Figure 4. Neutralization assay of convalescent sera. Sera from convalescent COVID-19 patients neutralized the SARS-CoV-2 pseudovirus. Serum sample from healthy individual was tested as a negative control. RLU were detected at 72 h post-pseudovirus inoculation. The data are presented as the mean percentages of inhibition ± SDs of triple wells. The neutralization titters (ID50) of the three convalescent sera are convalescent 1: 629, convalescent 2: 859 and convalescent 3: 1439, respectively.
Data analysis
All data are presented as means ± standard deviations (SDs). GraphPad Prism 8.0 software (GraphPad Software) was used to perform all statistical analyses and prepare graphs. Statistical significance was determined using one way ANOVA for multiple comparisons. Student’s t-tests were applied to compare the two groups. Differences with p-values < 0.05 were deemed statistically significant.
Acknowledgments
We would like to thank Prof. Cheguo Cai (Wuhan University, Wuhan, China) for providing the pNL4-3.Luc.R-E- plasmid. This work was supported by the Kuanren Talents Program of the second affiliated hospital of Chongqing Medical University, and the Emergency Project for Novel Coronavirus Pneumonia from the Chongqing Medical University (CQMUNCP0302).
This protocol was modified and improved from PMID: 32837985.
Competing interests
The authors declare no competing interests.
References
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 1995; 206: 935–944.
- Geraerts M, Willems S, Baekelandt V, et al (2006) Comparison of lentiviral vector titration methods. BMC Biotechnol 6:34. https://doi.org/10.1186/1472-6750-6-34
- Hu J, Gao Q, He C, Huang A, Tang N, Wang K. (2020). Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. Dec;7(4):551-557.
- Milone MC, O’Doherty U. Clinical use of lentiviral vectors. Leukemia 2018; 32: 1529–1541.
- Nguyen VP, Hogue BG. Protein interactions during coronavirus assembly. J Virol 1997; 71: 9278–9284.
- Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, Zhang L, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Fan C, Huang W, Xu M, Wang Y. (2020). Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. Dec;9(1):680-686
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. Mar 27;11(1):1620.
- Hu, J, Gao, Q, He, C, Huang, A, Tang, N and Wang, K(2021). Production and titration of SARS-CoV-2 spike pseudotyped virus using lentiviral system. Bio-protocol Preprint. bio-protocol.org/prep1303.
- Hu, J., Gao, Q., He, C., Huang, A., Tang, N. and Wang, K.(2020). Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes & Diseases 7(4). DOI: 10.1016/j.gendis.2020.07.006
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
