Advanced Search
Repair-Seq
Last updated date: Apr 12, 2021 Views: 1151 Forks: 0
IMPORTANT NOTE: PROTOCOL IS FOR ~500-750K CELLS
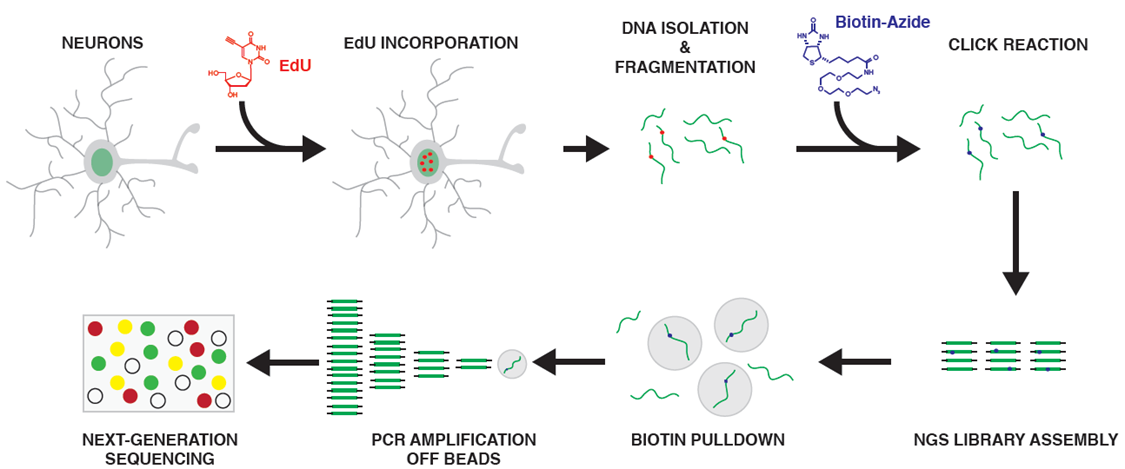
Isolate Genomic DNA (Dynabeads SILANE Genomic DNA Kit)
- Add 50 μL Proteinase K (20 mg/mL) to cell sample. Vortex 30 sec and then incubate for 2 min at room temperature.
- Add 350 μL Lysis/Binding Buffer (genomic DNA) to the sample and vortex for 15 sec to mix. Incubate for 5 min at 55°C and the vortex for 15 sec. Incubate again for a further 5 min at 55°C followed by a final vortex for 15 sec.
- Add 50 μL resuspended Dynabeads and vortex for 5 sec to mix. Add 400 μL isopropanol (100%). Vortex 30 sec and incubate on a roller for 3 min at room temperature followed by a final vortex for 30 sec.
- Plate the tube on the magnet and let the Dynabeads collect at the magnet for 2 min. Remove the supernatant by using a pipette. The Dynabeads/gDNA will form a nice pellet at the side of the tube, avoid touching the pellet with the pipette.
- Remove the magnet and add 950 μL Washing Buffer 1 (genomic DNA) vortex 30 sec at room temperature. The bead pellet should easily break into barely visible aggregates.
- Place the tube on the magnet for 2 min. Remove the supernatant.
- Repeat steps 5 and 6.
- Add 950 μL Washing Buffer 2 (genomic DNA). Vortex 30 sec at room temperature to resuspend the Dynabeads, and transfer the resuspended bead-solution to a clean tube.
- Plate the tube on the magnet for 1 min. Remove the supernatant.
- Add 950 μL Washing Buffer 2 (genomic DNA) and vortex 30 sec.
- Place the tube on the magnet for 1 min. Remove the supernatant. (Make sure that all the supernatant is completely removed in this last washing step. Use a small pipette tip and make sure that no droplets are left on the tube wall). Leave the tube on the magnet and let the beads pellet dry for 5 min at room temperature.
- Remove the magnet and add 100 μL Elution Buffer (genomic DNA). Completely resuspend the Dynabeads by vortexing and/or pipetting for 2 min at room temperature.
- Place the tube on the magnet and let the Dynabeads collect at the magnet for 1 min.
- Transfer the supernatant containing the purified genomic DNA to a clean tube.
DNA Fragmentation (Covaris m220)
(Protect gDNA from light)
15. Transfer genomic DNA (between 2-5 μg) to a Covaris MicroTUBE and adjust volume to 130 μL. Sonnicate with a Covaris m220 until fragments are between 350-450 bp for Sequencing.
Fragment with the following parameters:
Target BP (Peak) 450
Peak Incident Power (W) 50
Duty Factor 10%
Cycles per Burst 200
Treatment Time (s) 65
16. Check DNA fragmentation on Agilent TapeStation D1000 to ensure fragment size between 350-450bp.
17. Clean up gDNA fragments with AMPure XP beads (1:1.4 volume:volume). This is to remove the EDTA in the gDNA elution buffer:
- Add exactly 182 μL (1.4X volume) of beads to the reaction. Mix well by pipetting and incubate at room temperature for 10 minutes.
- Collect beads on a magnet and remove supernatant.
- Keeping the beads on the magnet, wash twice with 180 μL of 80% ethanol without mixing.
- Leave the beads on the magnet for 5 minutes to allow remaining ethanol to evaporate.
- To elute DNA, add 110 μL of ddH2O, gently mix by pipetting, incubate at room temperature for 10 minutes, separate on a magnet, and transfer the solution to a fresh 1.5 mL tube.
18. Collect 10 μL of sample for an INPUT control. Split remaining 100 μL into Clicked and “Unclicked/Negative” sample (omit biotin-TEG-azide, CuSO4, and Click-iT Buffer Additive in the negative reaction – replace with water).
Biotin-TEG-Azide Click Reaction for EdU
(Perform in the dark at room temperature)
19. Add the following components together sequentially from Life Technologies Click-iT EdU Imaging Kit (#C10337):
10 μL 10X Click-iT Buffer (10X TBS)
50 μL gDNA fragments
25 μL ddH2O
4 μL CuSO4
1 μL Biotin-TEG-Azide (0.1 mM final, Barry & Associates #BT1085)
10 μL Click-iT Buffer Additive
Incubate for 30 minutes at room temperature in the dark. Following the reaction clean with Ampure XP beads.
IMPORTANT NOTE: DO NOT ADD ANY OF THOSE TO THE UNCLICKED/NEGATIVE SAMPLE. JUST ADD 50 UL WATER AND PROCEED TO AMPURE CLEAN UP.
20. Add 140 μL (1.4X volume) of beads to the reaction. Mix well by pipetting and incubate at room temperature for 10 minutes.
21. Collect beads on a magnet and remove supernatant.
22. Keeping the beads on the magnet, wash twice with 180 μL of 80% ethanol without mixing.
23. Leave the beads on the magnet for 5 minutes to allow remaining ethanol to evaporate.
24. To elute DNA, add (22) μL of ddH2O, gently mix by pipetting, incubate at room temperature for 10 minutes, separate on a magnet, and transfer the solution to a fresh 1.5 mL tube. You may quantify DNA at this juncture with a NanoDrop spectrophotometer.
SwiftBio Accel-NGS 2S DNA Library Kit & Dual Index Kit
Performed according to manufacture specifications unless otherwise NOTED. Thaw all enzymes on ice for at least 10 minutes prior to making up Master mix.
Special Note: Each Library Reaction for the pull-down pull must use the same indexing. DNA input for reactions may range from 200-250 ng of input (1 day feed samples require 2 reactions minimum).
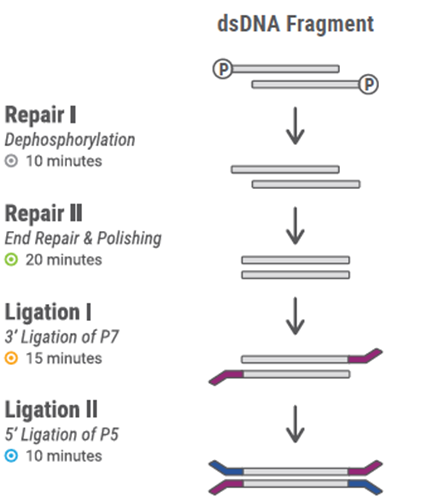
¤ Repair I
25. Transfer the fragmented dsDNA sample to a 0.2 mL PCR tube and adjust the volume of the sample to a final volume of 40 μL using Low EDTA TE, if necessary.
26. Add 20 μL of pre-mixed Repair I Master Mix (listed in the table below) to each sample containing the 40 μL DNA sample.
Reagents | Volume per Sample |
Low EDTA TE | 13 μL |
¤ Buffer W1 | 6 μL |
¤ Enzyme W2 | 1 μL |
Total Volume | 20 μL |
27. Mix by gently pipetting, place in the thermocycles, and run the Repair I Thermocycler Program in the order described below.
Sample Type | Thermocycler Program |
Repaired-Seq Inputs | 37°C, 10 min. lid heating OFF* |
*Alternatively, the thermocycler lid may be left open.
28. Clean up the Repair I reaction using a magnetic rack, Ampure XP Beads, and freshly prepared 80% ethanol. Follow the clean-up instructions below.
dsDNA Input | Insert Size | Sample Volume | Bead Volume (1.4X) | PEG NaCl Volume |
250-1000 ng gDNA fragments | 350-450 bp | 60 μL | 84 μL | - |
Section A: Size Selection/Clean-Up Protocol
Please use the following protocol for each clean-up step, substituting the correct Bead Volume, PEG NaCl Volume, and Elution Volume based on the table provided for each section.
- Ensure the magnetic beads are at room temperature and vortex beads to homogenize the suspension before use.
- Add the specified Bead volume or PEG NaCl volume to each sample. Mix by pipetting/vortexing. Quick spin the samples in a tabletop microcentrifuge.
- Incubate the samples for 5 minutes at room temperature.
- Place the sample on a magnetic rack until the solution clears and a pellet is formed (~2 minutes).
- Remove and discard the supernatant without disturbing the pellet (less than 5 μL may be left behind).
- Add 180 μL of freshly prepared 80% ethanol solution to the sample while it is still on the magnetic rack. Use care not to disturb the pellet. Incubate for 30 seconds and then carefully remove the ethanol solution.
- Repeat step 6 once more for a second wash with 80% ethanol solution.
- Quick spin the samples in a tabletop microcentrifuge and place back on the magnetic rack. Remove any residual ethanol solution from the bottom of the tube.
- Add the specified volume of each reaction mix (Repair II, Ligation I, and Ligation II) or elution volume (Post-Ligation II and Post-PCR library) of Low EDTA TE buffer and resusepnd the pellet. Mix well by pipetting up and down until homogenous.
29. Carefully remove and discard the supernatant without removing and magnetic beads.
¤ Repair II
30. Add 50 μL of pre-mixed Repair II Master Mix (listed in the table below to the beads for each sample and mix by pipetting until homogenous.
Reagents | Volume per Sample |
Low EDTA TE | 30 μL |
¤ Buffer G1 | 5 μL |
¤ Reagent G2 | 13 μL |
¤ Enzyme G3 | 1 μL |
¤ Enzyme G4 | 1 μL |
Total Volume | 50 μL |
31. Place the samples in the thermocycles, programmed at 20°C for 20 minutes with lid heating OFF.
32. Clean up the Repair II reaction using a magnetic rack, PEG NaCl solution, and freshly prepared 80% ethanol. Follow the clean up instructions below.
dsDNA Input | Insert Size | Sample Volume | Bead Volume | PEG NaCl Volume (1.2X) |
250-1000 ng gDNA fragments | 350-450 bp | 50 μL | - | 60 μL |
33. Carefully remove and discard the supernatant without removing any beads.
¤ Ligation I
34. Add 30 μL of pre-mixed Ligation I Master Mix (listed in teht able below) to the beads for each sample. Note: Reagent Y2, a truncated adapter, is provided separately in the Dual Indexing Adapter Kit.
Reagents | Volume per Sample |
Low EDTA TE | 20 μL |
¤ Reagent Y2 | 5 μL |
¤ Buffer Y1 | 3 μL |
¤ Enzyme Y3 | 2 μL |
Total Volume | 30 μL |
35. Place the samples in the thermocycler, programmed at 25°C for 15 minutes with lid heating OFF. Alternatively, the thermocycler lid may be left open.
36. Clean up the Ligation I reaction using a magnetic rack, PEG NaCl solution, and freshly prepared 80% ethanol. Follow the clean-up instructions below.
37.
dsDNA Input | Insert Size | Sample Volume | Bead Volume | PEG NaCl Volume (1.2X) |
250-1000 ng gDNA fragments | 350-450 bp | 30 μL | - | 36 μL |
¤ Ligation II
38. Add 50 μL of pre-mixed Ligation II Master Mix (listed in the table below) to the beads for each sample and reuspend by pipetting.
39.
Reagents | Volume per Sample |
Low EDTA TE | 30 μL |
¤ Buffer B1 | 5 μL |
¤ Reagent B2 | 2 μL |
¤ Reagent B3 | 9 μL |
¤ Enzyme B4 | 1 μL |
¤ Enzyme B5 | 2 μL |
¤ Enzyme B6 | 1 μL |
Total Volume | 50 μL |
Note: Reagent B2 (non-indexed adapter) is provided separately in the Indexed Adapter Kit.
40. Place the samples in the thermocycler, programmed at 40°C for 10 minutes with lid heating OFF (25°C hold). Alternatively, the thermocycler lid may be left open.
41. Clean up the Ligation II Reaction using a magnetic rack, PEG NaCl solution, and freshly prepared 80% ethanol. Follow the size selection instructions below.
dsDNA Input | Insert Size | Sample Volume | Bead Volume | PEG NaCl Volume (1.2X) |
250-1000 ng gDNA fragments | 350-450 bp | 50 μL | - | 60 μL |
42. At the end of the clean-up, resuspend beads in 20 μL of Low EDTA TE buffer.
43. the sample tubes on a magnetic rack and wait 2 minutes.
44. Carefully transfer the supernatant containing the final library to a clean tube without carrying any beads.
Biotin Pull-Down
(Perform all the following steps in 1.5 mL Eppendorf low-bind tubes)
45. Pool SwiftBio ChIP libraries from the same samples together following final elution from Ampure XP beads (40 uL).
46. Prepare for biotin pull-down by washing 10 μL of 10 mg/mL Dynabeads MyOne Streptavidin T1 beads (Life Technologies, 65602) with 600 μL of 1X Tween Washing Buffer (1X TWB). Separate on a magnet and discard the solution.
47. Resuspend the beads in 40 μL of 2X Binding Buffer (2X BB) and add to the SwiftBio ChIP library (40 uL). Incubate at room temperature for 30 minutes with rotation to bind biotinylated gDNA fragments to the streptavidin beads.
48. Separate on a magnet and discard the solution.
49. Wash the beads by adding 600 μL of 1X TWB and transferring the mixture to a new tube. Heat the tubes on a Thermomixer at 55°C for 2 min with mixing. Reclaim the beads using a magnet. Discard supernatant.
50. Repeat wash one more time.
51. Wash of beads in 100 μL of Low EDTA TE. Collect beads on a magnet and discard supernatant.
52. Resuspend in 20 μL of Low EDTA TE (you can freeze the beads at -20°C).
SwiftBio ChIP Library Amplification
53. Performed according to the Accel-NGS 2S DNA Library Kit & Dual Index Kit protocol using the 10 μL of beads as input.
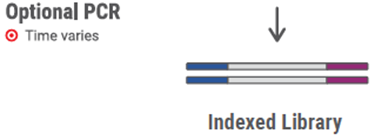
¤ PCR-Library Amplification
54. Add indexing reagent(s) directly to the entire eluted library (20 μL). Mix by pipetting. Note: Index D50X/D7XX, the indexed primers, are provided as part of the Dual Indexing Adapter Kit.
Reagents | Volume per Sample |
¤ Index D50X | 2.5 μL |
¤ Index D7XX | 2.5 μL |
Sample (T1 beads) | 20 μL |
Library + Primer Mix | 25 μL |
55. Add 25 μL of the pre-mixed Indexing PCR Master Mix (listed in the table below) to the entire eluted library (20 μL). Mix by pipetting.
Reagents | Volume per Sample |
Low EDTA TE | 10 μL |
¤ Reagent R2 | 4 μL |
¤ Buffer R3 | 10 μL |
¤ Enzyme R4 | 1 μL |
Total Volume | 25 μL |
56. Add 25 μL of the pre-mixed Indexing PCR Master Mix (listed in the table below) to the entire eluted library (20 μL). Mix by pipetting.
57. Amplify with the following Thermocycler Program
98°C for 3 minutes
PCR Cycles x15:
98°C for 10 seconds
60°C for 30 seconds
68°C for 60 seconds
68°C for 3 minutes
Hold at 4°C – proceed immediately to clean-up step.
58. Clean up the PCR Reaction using Ampure XP beads and freshly prepared 80% ethanol. Follow the size selection instructions below.
dsDNA Input | Insert Size | Sample Volume | Bead Volume (1.4X) | PEG NaCl Volume
|
250-1000 ng gDNA fragments | 350-450 bp | 50 μL | 70 μL
| - |
59. At the end of the clean-up, resuspend the beads in 20 μL of Low EDTA TE buffer.
60. Place the sample tubes on a magnetic rack and wait 2 minutes.
61. Carefully transfer the supernatant containing the final library to a clean tube without carrying any beads.
62. Check Repaired-Seq library on Agilent TapeStation with HS D1000 for library quality.
63. Perform size selection with Ampure XP beads
64. Quantify with Qbit prior to sequencing
65. Place samples at 4°C for short term or freeze at -20°C for longer term.
Reagents
SwiftBio Accel-NGS 2S Kit
SwiftBio Accel-NGS 2S Dual Indexing Kit
Swift PEG NaCl solution
Swift Low EDTA TE solution
2X BB (Binding Buffer)
10 mM TrisHCl pH 7.5
1 mM EDTA
2 M NaCl
1X TWB (Tween Wash Buffer)
5 mM Tris-HCl pH 7.5
0.5 mM EDTA
1 M NaCl
0.05% Tween-20
- Reid, D and Gage, F(2021). Repair-Seq. Bio-protocol Preprint. bio-protocol.org/prep1011.
- Reid, D. A., Reed, P. J., Schlachetzki, J. C. M., Nitulescu, I. I., Chou, G., Tsui, E. C., Jones, J. R., Chandran, S., Lu, A. T., McClain, C. A., Ooi, J. H., Wang, T., Lana, A. J., Linker, S. B., Ricciardulli, A. S., Lau, S., Schafer, S. T., Horvath, S., Dixon, J. R., Hah, N., Glass, C. K. and Gage, F. H.(2021). Incorporation of a nucleoside analog maps genome repair sites in postmitotic human neurons. Science 372(6537). DOI: 10.1126/science.abb9032
Category
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
