Advanced Search
Identification of Accessible Chromatin Regions with MNase-seq
*Contributed equally to this work Published: Mar 20, 2024 DOI: 10.21769/BioProtoc.4954 Views: 547
Reviewed by: Hassan Rasouli
Abstract
The study of accessible chromatin, also known as open chromatin, is currently a hot spot in the research of chromatin non-coding cis-regulatory elements and cis-trans controls of gene expression. Compared to animals, the accessible chromatin is different and relatively conserved across plant species. The identification of accessible chromatin regions (ACRs) in plants promotes our understanding of gene regulation, plant development, and regulatory changes underlying phenotypic evolution. Here, we describe an approach to identify wheat ACRs using differential MNase-seq. Micrococcal nuclease (MNase) is highly sensitive to digestion degree; it tends to cut accessible regions in case of light digestion and more closed regions in case of heavy digestion. We set up gradients of high- and low-concentration MNase digestion and performed high-throughput sequencing of DNA fragments near the length of mononucleosomes in the fragments digested by the two gradients. By comparing the differences in read enrichment under the two concentrations, we defined wheat genome regions highly sensitive to the change of digestion degree as ACRs and regions highly insensitive to the change as closed chromatin regions and identified nucleosome occupancy profiles as well. In short, we modified and refined the method from Rodgers-Melnick et al. (2016) for identifying open chromatin in maize, optimizing the nuclei extraction and ACRs identification for polyploidy, making its application in plants more intuitive, fast, and easy to operate. This method allows us to use MNase-seq to more easily identify ACRs in polyploid plants or large-genome species and to make multiple comparisons with ACRs obtained by other methods, so as to better facilitate the study of plant ACRs.
Graphical overview

Background
Accessible chromatin regions (ACRs) of eukaryotes generally imply the functional genome of the species or the collection of cis-regulatory elements (CREs) (Lu et al., 2019), such as regulatory elements located in promoters and enhancers (Yan et al., 2019). By identifying ACRs, important non-coding CREs and their roles in the regulation of gene expression, species growth and development, environmental adaptation, and natural evolution can be explored. There are several methods to identify ACRs, the most common of which are DNase-seq, ATAC-seq, and MNase-seq (Tsompana and Buck, 2014; Klein and Hainer, 2020). Unlike the enzymes DNase I used in DNase-seq and Tn5 used in ATAC-seq, MNase has both endonuclease and exonuclease activities, which digest and degrade naked DNA, leaving only sequences bound by repressors such as nucleosomes or DNA-binding proteins. The process is highly sensitive to the degree of digestion (i.e., MNase dose or concentration) and can be used to study nucleosome occupancy and digestion sensitivity, indirectly obtaining ACRs. Compared with DNase-seq and ATAC-seq, MNase-seq can identify some ACRs that cannot be identified by the other methods (Zhao et al., 2020). For example, in Arabidopsis thaliana, 20% more ARCs were identified by MNase-sensitive sites, obtained by sequencing 20–100 bp fragments, than by DNase-seq or ATAC-seq reads coverage. Meanwhile, MNase-seq can be used to estimate and contrast histone and non-histone DNA-binding components (Chereji et al., 2017) and identify both open and closed chromatin regions (Vera et al., 2014). However, the identification of plant ACRs with this method has also the disadvantage of demanding more sequencing material and higher sequencing depth, which requires weighing the pros and cons.
There are multiple strategies for identifying ACRs by MNase-seq. One is to select and sequence small DNA fragments (generally significantly smaller than the length of a mononucleosome, such as <80 bp or <100 bp), which may be the binding sites of some non-histone DNA-binding proteins (e.g., TF), directly based on MNase light digestion and define enrichment location of these small fragments as ACRs (Zhao et al., 2020). An alternative strategy is to define ACRs by processing with MNase digestion concentration gradients and sequencing DNA fragments that show a length close to that of mononucleosomes, to detect loci with changes in susceptibility to digestion (Rodgers-Melnick et al., 2016). Another strategy is to use MNase-seq and then combine it with ChIP-seq of histone subunit H3 or H4 for capture, to indirectly calculate the ACRs (Cook et al., 2017). Using these strategies, ACRs have been identified, and cis-acting elements have been found in human and mammalian species, Drosophila, yeast, Arabidopsis, maize, rice, and wheat (Fang et al., 2016; Mieczkowski et al., 2016; Rodgers-Melnick et al., 2016; Brahma and Henikoff, 2019; Schwartz et al., 2019; Jordan et al., 2020; Zhao et al., 2020). There may be differences in the ACRs obtained by the above strategies or in different species. This is mainly due to different research objectives, with differences in the definition of accessible chromatin and the target ACRs (a typical example is the controversy about fragile nucleosomes). Here, we provide a detailed protocol for this MNase-seq method in the identification of ACRs using the strategy of differential nuclease sensitivity, illustrated by data from polyploid crop wheat, while optimized for plant cell nuclei extraction and polyploid genome-specific alignment (Figure 1).
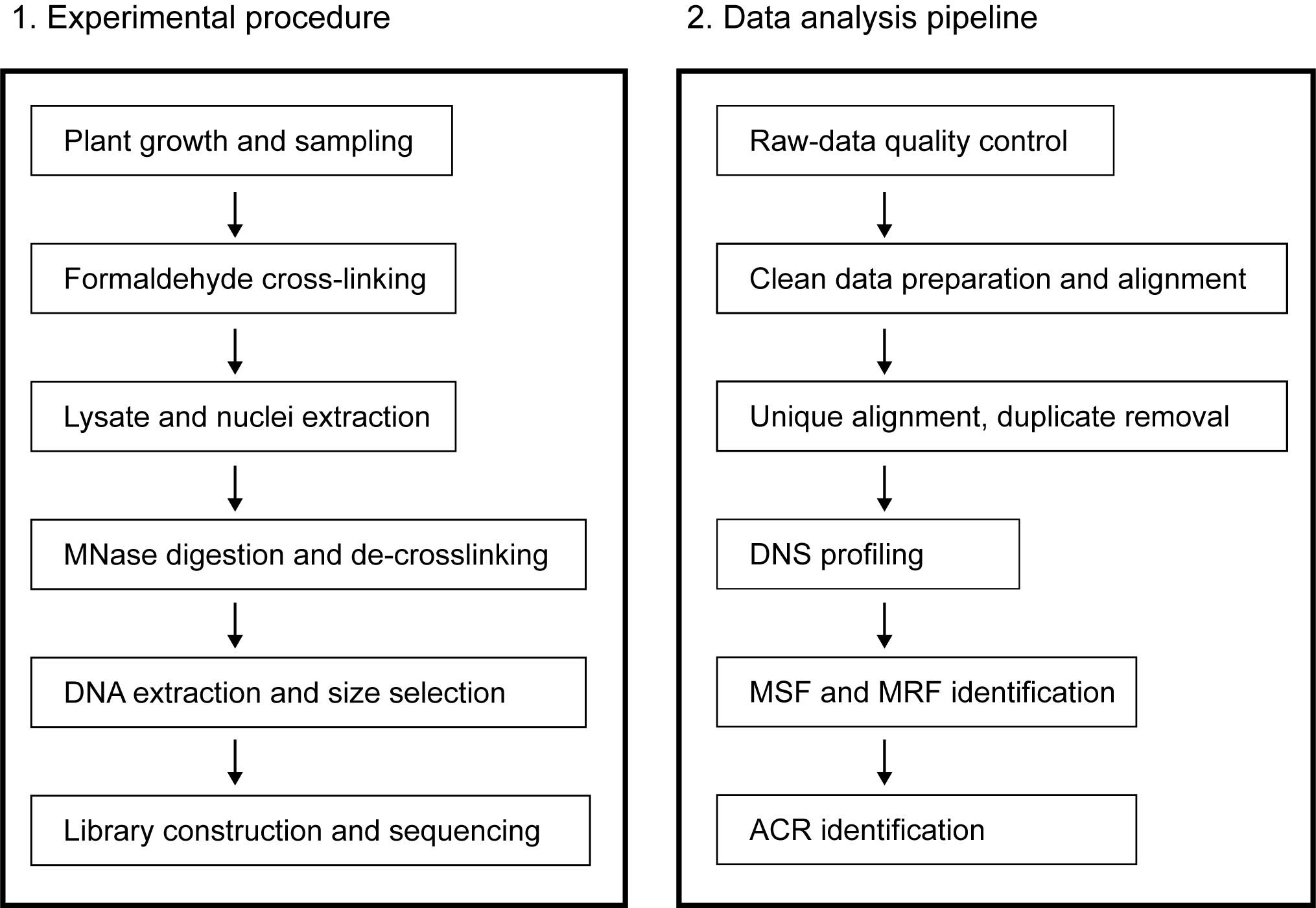
Figure 1. Schematic overview of the protocol.The experimental flow is shown on the left and the data analysis flow isshown on the right. MSF: MNase-sensitive footprint; MRF: MNase-resistantfootprint; ACR: accessible chromatin region.
Materials and reagents
Materials
Wheat seedlings at the three-leaf stage (Aikang58, CAAS)
Miracloth (Calbiochem, catalog number: 475855-1R)
50 mL centrifuge tube (Corning, catalog number: 430290)
5 mL pipette (Corning, catalog number: 4487)
PIPES (BBI, catalog number: A600719)
Sorbitol (BBI, catalog number: A610491)
EGTA (Millipore, catalog number: 324626)
DTT (Thermo Fisher, catalog number: R0861)
Spermine (Sigma-Aldrich, catalog number: S3256)
Spermidine (Sigma-Aldrich, catalog number: S2626)
37% Formaldehyde (Sigma-Aldrich, catalog number: 818708)
PMSF (Thermo Fisher, catalog number: 36978)
Glycine (Diamond, catalog number: A100167)
1 M Tris-HCl (pH 7.5) (Sangon, catalog number: B548124)
0.5 M EDTA (pH 8) (Sangon, catalog number: B540625)
Sucrose (Diamond, catalog number: A100335)
MgCl2 (Sigma-Aldrich, catalog number: M8266)
CaCl2 (Sigma-Aldrich, catalog number: C3306)
KCl (Sigma-Aldrich, catalog number: P9541)
NaCl (Sigma-Aldrich, catalog number: S3014)
SDS (Thermo Fisher, catalog number: 28364)
Phenol:Chloroform:Isoamyl alcohol 25:24:1 (Wako, catalog number: 311-90151)
RNase (Thermo Fisher, catalog number: EN0531)
Isopropanol (Sangon, catalog number: A507048)
1× TE buffer (Sangon, catalog number: B548106)
Glycerol (Diamond, catalog number: A100854)
Triton X-100 (Sigma-Aldrich, catalog number: T8787)
Percoll (GE, catalog number: 17-0891)
MNase (Thermo Fisher, catalog number: 88216)
Proteinase K (Solarbio, catalog number: 17-0891)
NEBNext UltraTM DNA Library Prep kit for Illumina (NEB, catalog number: E7645S)
Qiaex II gel extraction kit (Qiagen, catalog number: 20021)
Solutions
Nuclei isolation buffer (see Recipes)
Fixation buffer (see Recipes)
Percoll cushion solution (see Recipes)
MNase digestion buffer (see Recipes)
Recipes
Note: All concentrations listed are final concentrations.
Nuclei isolation buffer
15 mM PIPES (NaOH at pH 6.8)
0.32 M sorbitol
80 mM KCl
20 mM NaCl
0.5 mM EGTA
2 mM EDTA
1 mM DTT
0.15 mM spermine
0.5 mM spermidine
Fixation buffer
Nuclei isolation buffer, add 1% formaldehyde and 0.1 mM PMSF freshly
Percoll cushion solution
50% (vol/vol) Percoll in nuclei isolation buffer
MNase digestion buffer
50 mM Tris-HCl at pH 7.5
320 mM sucrose
4 mM MgCl2
1 mM CaCl2
Equipment
Mortar and pestle (Avantor, catalog number: HALDL55/1/G)
Beaker (30 mL) (PYREX, catalog number: 1000-30)
Magnetic stirrer (Thermo Fisher, catalog number: S194615)
Hybridization oven (Galanz, catalog number: P70J17L-V1)
4 °C centrifuge (Eppendorf, catalog number: 5804)
Nanodrop 2000 (Thermo Fisher, catalog number: 13-400-412)
Software
Deeptools v3.4.3 (Max Planck Institute for Immunobiology and Epigenetics; https://deeptools.readthedocs.io/en/latest/index.html) (February 2022)
Samtools v1.6 (Genome Research Limited; http://www.htslib.org/doc/samtools.html) (February 2022)
BEDTools v2.29.1 (University of Utah; https://bedtools.readthedocs.io/en/latest/content/bedtools-suite.html) (February 2022)
R v3.2.2 (Lucent Technologies; https://www.r-project.org/) (March 2022)
IGV v2.5.0 (University of California; http://software.broadinstitute.org/software/igv/) (March 2022)
Trimmomatic v0.36 (THE USADEL LAB; http://www.usadellab.org/cms/?page=trimmomatic) (February 2022)
Bowtie2 v2.3.4 (Johns Hopkins University; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) (February 2022)
FastQC v0.11.8 (Babraham Bioinformatics; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (February 2022)
Procedure
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
Category
Plant Science > Plant molecular biology > DNA > DNA sequencing
Molecular Biology > DNA > Chromatin accessibility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
