Advanced Search
Identification of Topologically Associating Domains (TADs) and Long-distance Enhancer–gene/gene–gene Interactions with Hi-C and HiChIP
Published: Nov 20, 2023 DOI: 10.21769/BioProtoc.4879 Views: 510
Abstract
The hierarchy featured architecture of 3D genome influences the expression of genes to regulate development and defense processes. Hi-C is one of the most widespread technologies to explore hierarchy 3D genome organizations, including compartment, topologically associating domains (TADs), and loops. HiChIP can accurately identify chromatin loops between cis-regulatory elements in non-coding regions and genes. At the same time, growing innovative methods can be used to identify different hierarchy architectures of 3D genome, but most analyses are performed with a combination of a variety of software. Therefore, a clear and complete pipeline is essential. We summarize the detailed usage of various tools for identifying the 3D genome structure using Hi-C and HiChIP, as well as for visualizing the final result. Using these software tools, we identify the TADs and loops from the Hi-C and HiChIP libraries in human cell lines and cotton plants. In brief, this pipeline will help researchers choose a suitable tool with less time cost.
Keywords: 3D genomeBackground
The genetic diversity of organisms is the fundamental basis for forming complex and diverse global ecosystems. Adequate and comprehensive knowledge of genetic information will help us understand the nature of evolution more deeply and thus improve our living environment without affecting the survival of other species. The study of the interpretation of genetic information from multiple perspectives has been booming (Misteli, 2020; Purugganan and Jackson, 2021; Song et al., 2021; Zhang et al., 2021). Among them, the 3D genome has become a fascinating study field. It has been extensively used to comprehensively study how the hierarchy of 3D genome regulates biological processes such as evolution, development, and resistance to pathogens by combining 1D genomics, transcriptomics, and epigenomics (Fullwood et al., 2009; Lieberman-Aiden et al., 2009; Mumbach et al., 2017; Sun et al., 2018; Wang et al., 2018; Norrie et al., 2019; Zheng and Xie, 2019; Yang et al., 2022). So far, there is a growing body of literature on topologically associating domains (TADs) and loops, since they are finely architected to regulate changes in genetic information (Rao et al., 2014; Wang et al., 2018; Zheng et al., 2019; Espinola et al., 2021; Hoencamp, 2021). Hence, accurate identification of TAD and loop structures is essential for understanding how the hierarchy of 3D genome architecture regulates biological processes.
With the innovation in experimental methods of 3D genomic study and the advancement of sequencing technologies, a large number of software tools were developed to identify 3D genome structures (Ay et al., 2014; Wang et al., 2015; Forcato et al., 2017; Bhattacharyya et al., 2019; Yardımcı et al., 2019; Fernandez et al., 2020; Wolff et al., 2020; Mourad, 2022). They have contributed to a boom in the field of 3D genomic research. This also increases the difficulties for researchers to realize them (Forcato et al., 2017). Therefore, a complete order pipeline is very useful for scientific researchers. To achieve this, we summarized the processing of several software tools, including identifying TADs by TADLib and Juicer, inferring loops of Hi-C data by Fit-Hi-C and Juicer, and inferring loops of HiChIP data by FitHiChIP and hichipper. The integration of these software into a whole pipeline by combining HiC-Pro and Juicebox can provide final visualization results from raw reads (Figure 1). The pipeline provides a comprehensive 3D genome analysis process that is applicable not only to Hi-C datasets but also compatible with HiChIP datasets. Moreover, the pipeline eliminates the need for complex intermediate steps in multiple software, providing users a simple and efficient experience while preserving the software's customizable function. In summary, the pipeline provides user-friendly features that are especially beneficial for researchers who are new to 3D genome analysis.
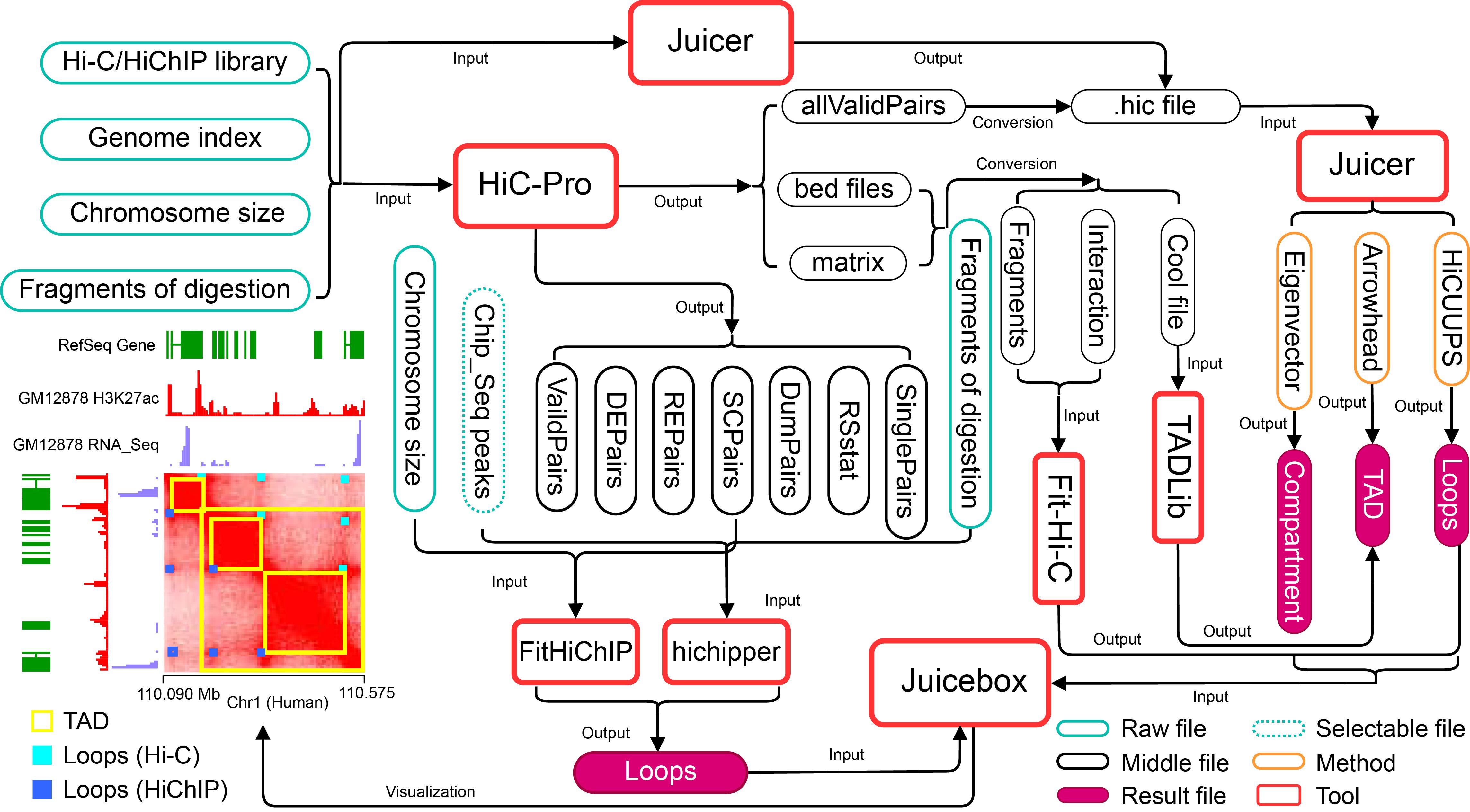
Figure 1. Flowchart for the identification of topologically associating domains (TAD) and loops from the datasets of Hi-C and HiChIP by different software
Software and datasets
Software
Python (Version 3.9.5/Version 2.7.18, https://www.python.org/downloads/) (2020/09)
R (Version 4.0.0, https://cran.r-project.org/bin/windows/base/old/) (2020/09)
Bowtie2 (Version 2.4.4) (Langmead and Salzberg, 2012)
BWA (Version 0.7.17) (Li and Durbin, 2009)
SAMtools (Version 1.9) (Li et al., 2009)
BEDTools (Version 2.27) (Quinlan and Hall, 2010)
MACS2 (Version 2.1.1) (Zhang et al., 2008)
HiC-Pro (Version 2.11.4) (Servant et al., 2015)
Juicer (Version 1.6) (Durand et al., 2016)
Juicer tools jar (Version 1.22.01, https://github.com/aidenlab/juicer/wiki/Download) (2020/11)
Juicebox (Version 1.11.08, https://github.com/aidenlab/Juicebox/wiki/Download) (2020/11)
TADLib (Version 0.4.1) (Wang et al., 2015)
HiCPeaks (Version 0.3.4, https://github.com/XiaoTaoWang/HiCPeaks) (2021/04)
Fit-Hi-C (Version 2.0.8) (Ay et al., 2014)
FitHiChIP (Version 9.1) (Bhattacharyya et al., 2019)
hichipper (Version 0.7.7) (Lareau and Aryee, 2018)
Data
Library of Hi-C
Hi-C data of human GM12878 B-lymphoblastoid cells (Rao et al., 2014)
Hi-C data of cotton fiber of Gossypium barbadense 3–79 at 20 days post anthesis (DPA) (Pei et al., 2022)
Library of HiChIP
HiChIP data (H3K27ac) of human GM12878 B-lymphoblastoid cells (Mumbach et al., 2017)
Procedure
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
Category
Bioinformatics and Computational Biology
Systems Biology > 3D Genomics
Systems Biology > Genomics
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

