Advanced Search
Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells
*Contributed equally to this work Published: Vol 11, Iss 5, Mar 5, 2021 DOI: 10.21769/BioProtoc.3936 Views: 7719
Reviewed by: Vikash VermaAndrew L. EagleYoshihiro Adachi
Abstract
Monitoring vesicle trafficking is an excellent tool for the evaluation of protein dynamics in living cells. Such study is key for the understanding of protein sorting and secretion. Recent developments in microscopy, as well as new methodologies developed to study synchronized trafficking of proteins, allowed a better understanding of signaling, regulation and trafficking dynamics at the secretory pathway. One of the most helpful tools so far developed is the Retention Using Selective Hooks (RUSH) system, a methodology that facilitates the evaluation of synchronized cargo trafficking by monitoring fluorescent vesicles in cells upon biotin addition. Here we present a protocol that allows the quantitative evaluation of protein cargo trafficking at different fixed time points and an analytic approach that enables a better examination of specific cargo trafficking dynamics at the secretory pathway.
Graphic abstract:
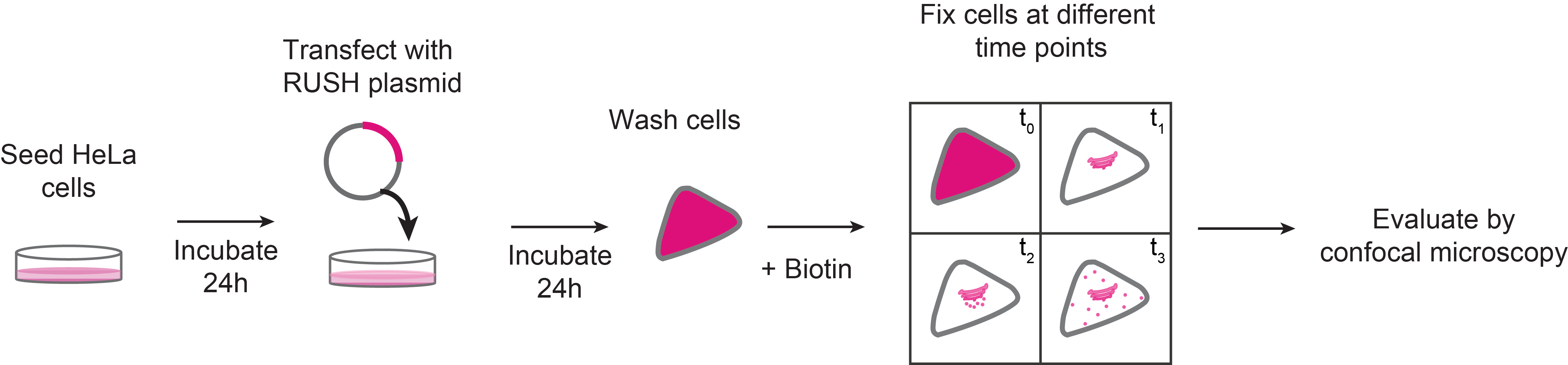
Schematic representation of RUSH sorting assay in mammalian cells
Background
Monitoring vesicle trafficking is an excellent tool for the evaluation of secretory protein dynamics in living cells. Given that around 30% of the total amount of newly synthesized proteins in mammals follow the secretory pathway (Pfeffer, 2010; Boncompain and Weigel, 2018), the study of its trafficking dynamics is key for the understanding of protein sorting and secretion. Proteins that follow the secretory pathway go through several maturation steps, starting at the ER, they are transported along the different Golgi stacks until they reach the trans-Golgi network (TGN), a sorting station. In the TGN they are finally packed into vesicles and initiate their route to their final destination (Glick and Luini, 2011; Pantazopoulou and Glick, 2019).
Recent developments in microscopy, as well as new methodologies developed to study the synchronized trafficking of proteins allowed to better understand signaling, regulation and trafficking dynamics at the secretory pathway (Stephens and Perez, 2013). One of the most helpful tools so far developed is the Retention Using Selective Hooks (RUSH) system, a methodology that facilitates the evaluation of synchronized cargo trafficking by monitoring fluorescent vesicles in cells upon biotin addition (Boncompain et al., 2012).
The RUSH system has two main elements: a protein of interest (POI) tagged with a fluorophore and bound to a streptavidin binding peptide (SBP); and a streptavidin molecule bound to a retention signal (known as the “hook”, e.g., a KDEL sequence for retention in the ER; Figure 1). This complex, in the absence of biotin will be retained in the donor compartment, but once biotin is added to the culture medium, the POI-fluorophore-SBP complex will be released to the next compartment. This set-up allows to follow up the POI at different time points by confocal microscopy (Boncompain et al., 2012).
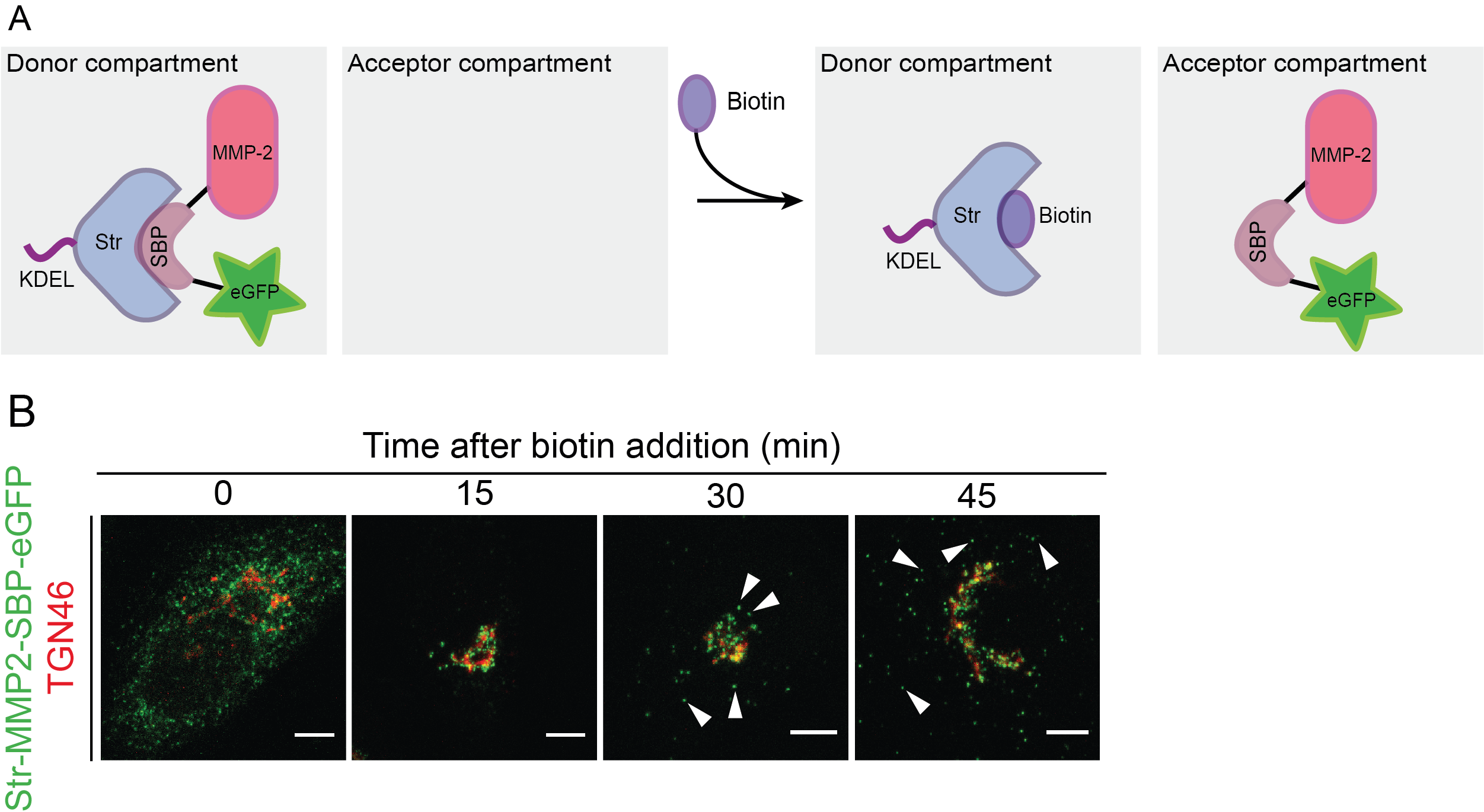
Figure 1. Scheme representing the RUSH system. A. Protein complex containing the protein of interest (here matrix metalloprotease 2, MMP2), a streptavidin binding peptide (SBP) and a fluorescent protein (eGFP). The complex is bound via SBP to streptavidin (Str), which is linked to a KDEL sequence for its retention in the endoplasmic reticulum (donor compartment). Without biotin addition, the complex remains in the donor compartment; however, once biotin is added to the media, it binds to streptavidin and enables the trafficking of the MMP2-SBP-eGFP protein complex to the acceptor compartment. Figure adapted from Boncompain et al. (2012). B. Immunofluorescence images showing the trafficking of MMP2 across the secretory pathway, with MMP2 localizing at the ER when biotin is absent, and later localizing at the Golgi (15 and 30 min) and in post-Golgi vesicles (45 min) upon biotin addition. TGN46: trans-Golgi marker. Figure taken from Pacheco-Fernandez et al. (2020).
Among other techniques, the RUSH system has the advantage of enabling the monitoring of cells under physiological conditions. Given that biotin is a non-toxic small molecule, the synchronization of cargo release does not lead to high stress for the cells (Boncompain and Perez, 2013). Also, compared to other techniques such as the light-triggered protein secretion system (Chen et al., 2013), it does not require a previous aggregation of the protein, which may lead to trafficking through different routes, higher protein degradation rates, and consequently, higher cell toxicity (Boncompain and Perez, 2013).
Building on the original RUSH methodology, here we present a protocol that allows to quantitatively evaluate protein cargo trafficking at different fixed time points. Importantly, in this protocol we provide a quantitative analysis to better examine intra-Golgi trafficking dynamics by evaluating specific cargo trafficking dynamics along the secretory pathway using confocal microscopy. This allows to evaluate the impact of knocking-out or silencing secretory pathway proteins on cargo trafficking (Crevenna et al., 2016; Deng et al., 2018; Pacheco-Fernandez et al., 2020).
Materials and Reagents
Circle coverslips, thickness 1 (ThermoScientificTM, catalog number: CB00120RA120MNT0 )
6-well cell culture plates (Corning®, Costar®, catalog number: CLS3516-1EA )
SuperfrostTM Microscopy Slides (ThermoScientificTM, catalog number: AA00008332E00MNT10 )
Polyethylenimine, linear, MW 25,000, transfection grade (PEI 25KTM; Polysciences, catalog number: 23966-1).
60-70% confluent HeLa cells
1× Dulbecco’s Phosphate Buffered Saline (DPBS), no calcium, no magnesium (Life Technologies, Gibco, catalog number: 14190144 ). Storage temperature: 4 °C.
pIRESneo3-Str-KDEL-MMP2-SBP-EGFP vector (Figure 2). Storage temperature: -20 °C
Important note: This plasmid was generated by replacing the ST (ST6GAL1) sequence by MMP2 (our protein of interest) in the Addgene plasmid number 65264 (Str-KDEL_ST-SBP-EGFP).
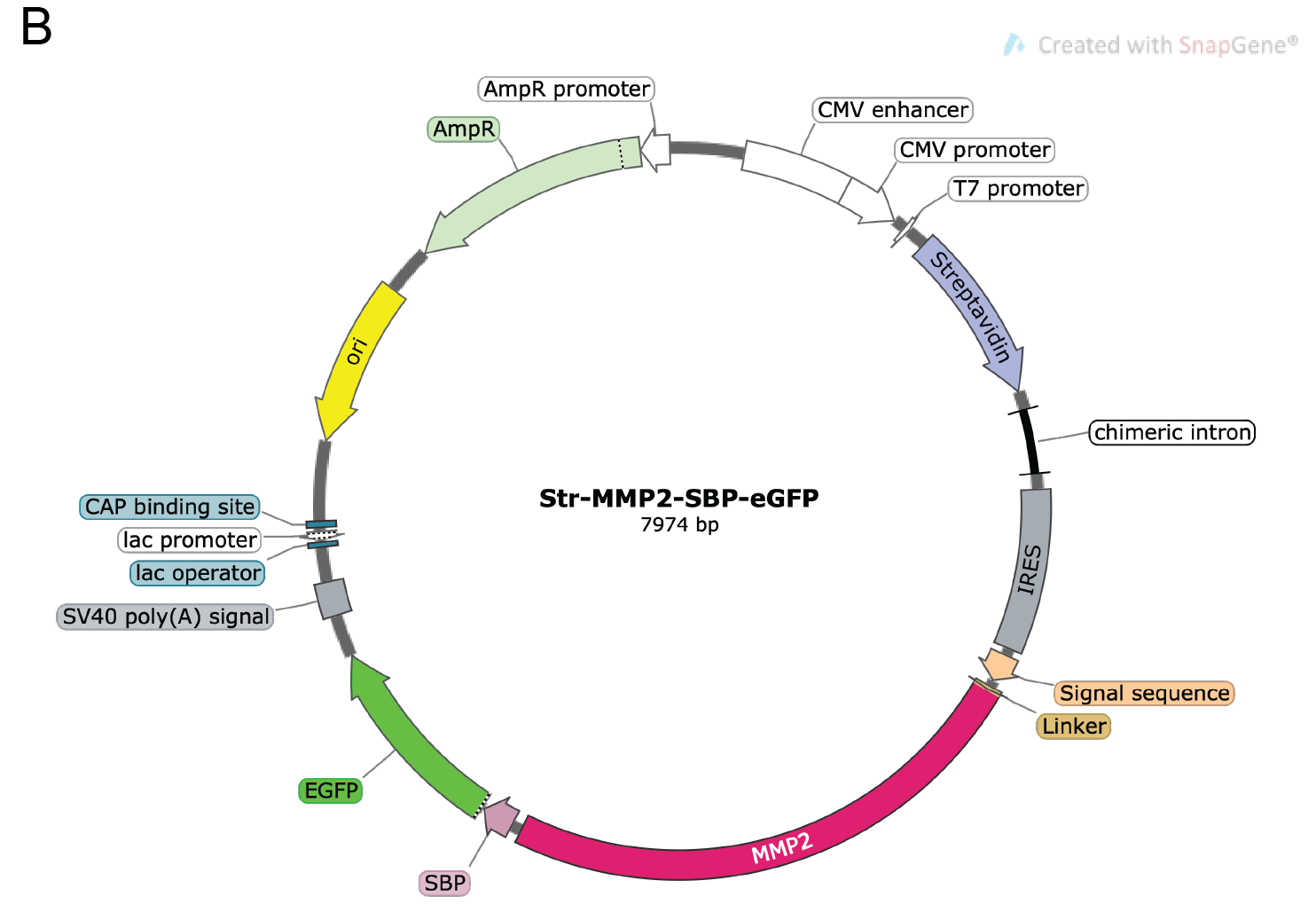
Figure 2. Vector map for the pIRESneo2-Str-KDEL-MMP2-SBP-eGFP construct used as an example in this protocol. The plasmid depicts the KDEL-bound streptavidin and the reporter MMP2-SBP-eGFP. An IRES element in the plasmid enables the simultaneous expression of both Str-KDEL and MMP2-SBP-eGFP in one single plasmid. For more details about the construct, please see Boncompain et al. (2012). The map was generated with SnapGene®.Opti-MEM® reduced serum media (Life Technologies, Gibco, catalog number: 31985070 ). Storage temperature: 4 °C
500 mM d-biotin (Merck, SUPELCO, catalog number: 47868 ). Storage temperature: 4 °C
4% paraformaldehyde (PFA) in 1× PBS. Prepared using the formaldehyde solution, ROTIPURAN® 37%, p.a., ACS (Roth, catalog number: 4979.1 )
Primary antibodies
For the protein of interest
Depending on the analysis, specific organelle markers can be used. For example, calnexin can be used as an ER marker, whereas, depending on the analytical evaluation, different Golgi markers could be used (e.g., GM-130 for cis-Golgi and TGN-46 for trans-Golgi network)
Secondary antibodies
Our studies were performed with Alexa-Fluor antibodies 488, 594 and 633 targeting different species (Life Technologies, ThermoScientific, catalog number varies according to species and wavelength). Storage temperature: 4 °C.
ProLong Gold antifade reagent (Life Technologies, Invitrogen, catalog number: P36934 )
PEI solution at 1 mg/ml (see Recipes)
Dulbecco’s Modified Eagles Medium (DMEM) complete medium (see Recipes)
Permeabilizing solution (see Recipes)
Triton X-100 (Carl Roth, catalog number: 3051.2 )
Sodium dodecyl sulphate (SDS) 10% solution
Blocking solution (see Recipes)
Bovine Serum Albumin (BSA) lyophilized powder (PAN Biotech, catalog number: P06-1391050 )
Phosphate buffered saline (PBS) powder (Sigma, catalog number: P3813-1PAK ): Prepare a 1× solution according to manufacturer’s instructions
Equipment
Tweezers
Laminar flow hood
Cell incubator set at 37 °C, 5% CO2
Water bath set at 37 °C
Zeiss laser scanning LSM780 confocal microscope (Carl Zeiss)
Zeiss 100× (NA 1.46, oil) objective (Carl Zeiss)
488-nm laser line
Software
Zeiss Zen software 2010 (Zeiss, https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html)
ImageJ v.1.37 (National Institutes of Health [NIH], https://imagej.nih.gov/ij/)
ImageJ macro for “RUSH-Vesicle-Analysis” (https://github.com/MehrshadPakdel/RUSH-Vesicle-Analysis)
Procedure
Category
Cell Biology > Cell imaging > Confocal microscopy
Biochemistry > Protein > Self-assembly
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link