Advanced Search
Detecting Spaciotemporal Transcript Accumulation in Maize by RNA In Situ Hybridization
*Contributed equally to this work Published: Feb 20, 2021 DOI: 10.21769/BioProtoc.3931 Views: 3102
Edited by: Marisa Rosa
Abstract
RNA in situ hybridization is a method for visualizing spatiotemporal transcript accumulation in cells and tissues. The method provides clear resolution, is highly sensitive and specific, and can uncover gradients of transcript accumulation within a histologically-intact tissue, which is not possible currently with other methods for transcript detection. RNA in situ hybridization, however, is not a quantitative approach for gene expression. Protocols for RNA in situ hybridization have numerous steps that can span several days of work, complicating troubleshooting procedures. Here, we build on previously published RNA in situ hybridization protocols optimized for paraffin-embedded and sectioned maize tissue (Jackson, 1991; Long et al., 1996 ; Javelle et al., 2011) by providing additional measures for optimized transcript detection.
Graphic abstract: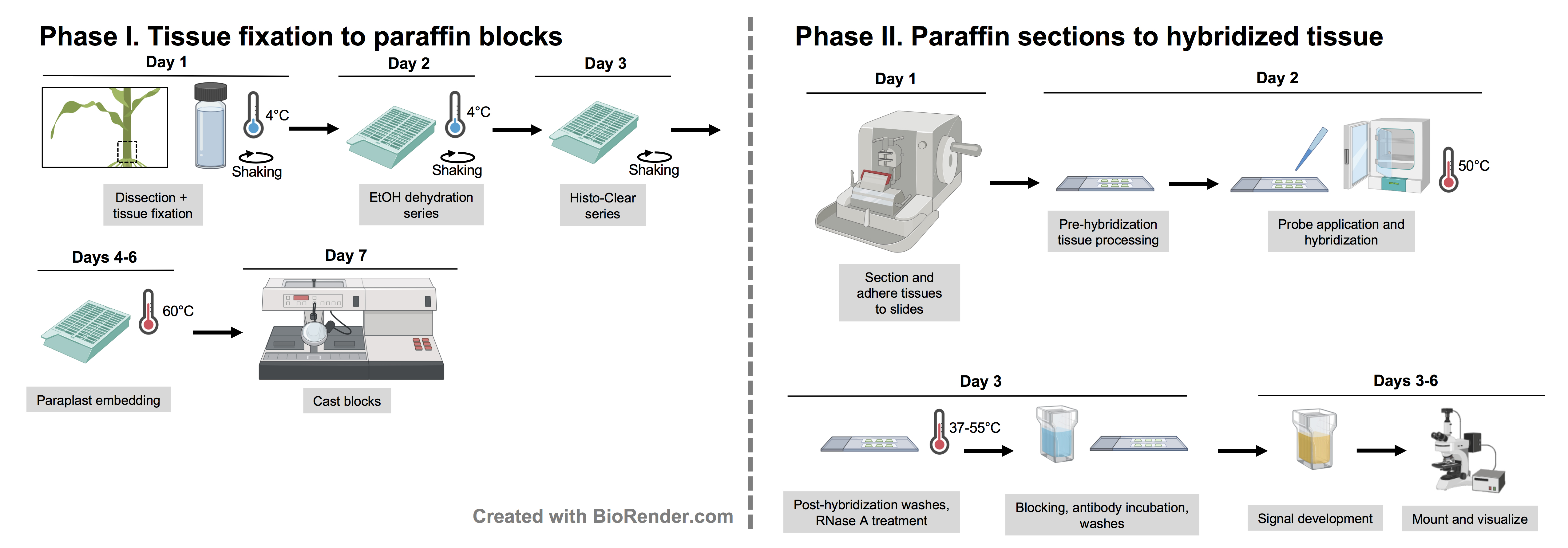
Workflow for RNA in situ hybridization
Background
Differential gene expression in time and space allows cells possessing the same genetic material to take on different identities. Indeed, such shifts in gene expression are often responsible for driving evolutionary changes in patterning or morphology across organisms (Carroll, 2005). Therefore, insights into developmental processes are enhanced by observations of gene expression in its native context within a tissue. Approaches such as RT-qPCR and RNA-Seq rely on the destructive processing of tissues to extract RNA for gene expression analysis. RNA in situ hybridization offers the advantage of providing spatially and temporally resolved analyses of gene expression within the native tissue and cell type context. In maize and other plant species, RNA in situ hybridization has been used to probe gene function, tissue patterning, the evolutionary modification of gene expression, and corroborate single-cell expression data (Jackson et al.,1994 ; Whipple et al., 2010; Johnston et al., 2014; Strable et al., 2017; Knauer et al., 2019 ; Satterlee et al., 2020). Here, we present a method for detecting the accumulation of transcripts by RNA in situ hybridization in maize, a model genetic system and important staple crop. This protocol demonstrates the technique in shoot apex tissue sections where new leaf and stem tissue is initiated from pluripotent cell populations within the shoot apical meristem. Hybridization of an antisense antigen-tagged RNA probe to complementary RNAs within the tissue enables the qualitative detection of transcript accumulation patterns by an enzyme-conjugated antibody that catalyzes a colorimetric staining reaction. This protocol is readily adaptable to other maize tissues and is based on Jackson (1991) and Javelle and coworkers (2011).
Materials and Reagents
Consumables
Paper towels
Kimwipes
Razor blades (single edge, steel)
Pencil
Parafilm
Adhesive Glass Microscope Slides (VWR, VistaVisionTM HistoBond®, catalog number: 16004-406 )
Cover slips (25 × 60 mm; VWR, catalog number: 89082-272 )
Glass Pasteur pipettes
Glass vials with caps (VWR, catalog number: 66012-022 )
Paraplast Plus® (McCormick Scientific, catalog number: 15159-464 )
Cups with lids (VWR, catalog number: 89508-714 )
Biopsy cassettes (VWR, catalog number: 25608-756 )
Disposable base molds (VWR, catalog number: 100501-996 )
Embedding ring (VWR, catalog number: 87002-374 )
Biological material
Maize tissue (e.g., shoot apex, inflorescence, root)
Reagents
Nuclease free water
Ethanol (VWR, catalog number: EM-EX0276-3S )
Histo-Clear II (VWR, catalog number: 101412-882 )
Acetic acid glacial (VWR, catalog number: BDH3094-2.5LG )
37% Formaldehyde solution (VWR, catalog number: 97064-604 )
Permount (Fisher, catalog number: SP15-100 )
Ethanol series: dilutions to 95%, 85, 70, 50 in distilled water
Eosin Y (VWR, catalog number: 97061-034 )
TOPOTM Cloning Kit for Sequencing (Thermo Fisher Scientific, catalog number: K4575-01) or pGEM®-T Easy Vector System (Promega, catalog number: A1380 )
RNase OUT; 40 U/μl (ThermoFisher, catalog number: 10777019 )
RQ1 RNase-Free DNase; 1 U/μl (Promega, catalog number: M6101 )
DIG RNA labeling mix (Roche, catalog number: 11277073910 )
SP6 RNA polymerase 1,000 U (Roche, catalog number: 10810274001 )
T3 RNA polymerase 1,000 U (Roche, catalog number: 11031163001 )
T7 RNA polymerase 1,000 U (Roche, catalog number: 10881767001 )
RNase-A (Roche, catalog number: 10109142001 )
Blocking Reagent (Roche, catalog number: 11096176001 )
Anti-Digoxygenin-AP, Fab fragments from sheep, 150 U (Roche, catalog number: 11093274910 )
NBT/BCIP stock solution (Roche, catalog number: 11681451001 )
Mini Quick Spin RNA columns (Sigma, catalog number: 11814397001 )
QIAprep Spin Miniprep Kit (Qiagen, catalog number: 27104 )
DNA clean & concentrator kit (Zymo Research, catalog number: D4033 )
tRNA from E. coli (Roche, catalog number: 10109541001 )
Triton® X-100 (Sigma, catalog number: X100 )
Deionized formamide (Sigma, catalog number: S4117 )
Protease from Streptomyces griseus Type XIV (Sigma, catalog number: P5147 )
Triethanolamine-HCl (Sigma, catalog number: T1502 )
Acetic anhydride (Sigma, catalog number: 320102 )
Dextran sulfate 50% solution (Sigma, catalog number: S4030 )
Denhardt’s solution 50× concentrate (ThermoFisher, catalog number: 750018 )
Albumin from bovine serum ≥98% (Sigma, catalog number: A9418 )
TAE-agarose gel
Glycine
1 M Tris-HCl (at pH 7.5, 8.0, and 9.5)
0.5 M EDTA
5 M NaCl
FAA fixative (see Recipes)
1× PBS buffer (see Recipes)
2× carbonate buffer (see Recipes)
10% Glacial Acetic Acid (see Recipes)
3 M NaOAc pH 5.2 (see Recipes)
4 M NH4OAc (see Recipes)
1× TBS buffer (see Recipes)
1× TN buffer (see Recipes)
Protease buffer (see Recipes)
1× in situ hybridization salts (see Recipes)
0.2× SSC (see Recipes)
1× NTE buffer (see Recipes)
Equipment
Pipettes
Forceps (VWR, catalog number: 82027-386)
Slide warmer (set to 37-42 °C)
Oven set at 42 °C
Vacuum pump
Microtome
Fine paintbrushes (e.g., Virtuous Arts Fine Paintbrushes, amazon.com)
Stainless steel slide rack with handle (VWR, catalog number: 25461-014 )
Tupperware (e.g., medium rectangular 15.5 × 8.1 × 7.3 cm of volume 412 ml)
Large Tupperware with sealable lids
Glass staining dish (VWR, catalog number: 25461-016 ) with cover (VWR, catalog number: 25461-018 )
Orbital shaker
Heating block
Incubator up to 60 °C capacity
Ovens and water baths for 37 °C and 55 °C
Centrifuge at room temp and 4 °C
Fume hood
Gel electrophoresis setup
Microscope equipped with a camera
Procedure
Category
Plant Science > Plant molecular biology > RNA > RNA detection
Developmental Biology > Morphogenesis > Organogenesis
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
