Advanced Search
An Efficient, Low-cost and Scalable Method for Constructing a Thousand of Synthetic Communities (SynComs)
Published: Sep 5, 2024 DOI: 10.21769/BioProtoc.2405576 Views: 375
Abstract
Recent advances in the study of complex microbial communities have highlighted the necessity of large-scale development and application of synthetic communities (SynComs) to investigate microbiome functionality and ecology within various fields such as agriculture, medicine, and environmental sciences. Despite the exponential increase in potential combinations within SynComs correlating with the number of involved strains (totaling 2N combinations for N strains), research has often focused on a limited subset of possible combinations due to these constraints. This limitation could potentially impact the conclusions drawn about mechanisms of emergent functionsy within microbial communities. In order to exhaustively construct SynComs from multiple species, this paper integrates the permutation and combination method with commonly used laboratory materials such as microtiter plates and pipettes, and develops a program to perform tasks like unique microbial community id assignment and data collection form generation. This simplifies the process of manually constructing over 1000 different SynComs in the laboratory. Compared to conventional methods, this method is more efficient, cost-effective, and scalable, and it avoids the sampling process confusion that can occur with conventional methods. Herein, we provide detailed experimental procedures for constructing 16-2048 different SynComs using 4-11 species as examples. Moreover, the program ‘syncons’ is provided as an R package that can be operated locally or accessed via cloud service, offering functionalities such as procedure overview, SynComs id assignment, and data collection form generation. Given the significant potential of SynComs, this method can serve as a fundamental experimental operation in research topics such as microbial co-cultivation and multispecies biofilms, offering broad application value.
Keywords: SynComsBackground
Advancements in the investigation of complex microbial community structures and functions have rendered the development and utilization of synthetic communities (SynComs) [1], which are essentially employed for exploring microbiome functionality and microbial ecology across diverse sectors, including agriculture [2], medicine [3], ecology [4], environmental science [5], and energy [6]. Future applications in designing SynComs are anticipated to grow, potentially involving combinations of dozens or hundreds of strains7. Theoretically, possible combinations within SynComs rise exponentially as a function of the number of strains used, for example, a system with 10 strains yields 1,024 possible different combinations is sum. This combinatorial explosion has historically confined research to only a fraction of potential combinations, and this may potentially limit understanding of the emergence and dynamics of microbial communities. To meet specific research needs, it is sometimes necessary to exhaustively construct all possible combinations of synthetic microbial communities.
Currently, the construction of many different SynComs is primarily accomplished using methods such as manual assembly, automated devices, and microdroplet technologies. Among these, automated equipment, such as the apricot DC1 pipetting workstation, has achieved a degree of high repeatability and precision, but at the expense of some flexibility and at a cost not all laboratories can afford. Furthermore, while microdroplet technology has shown immense potential in high-throughput screening of SynComs, its application is currently limited by numerous technical challenges8. Following traditional construction paradigms, manual assembly is undoubtedly the least efficient and most error-prone approach. However, this method, through judicious planning of the addition sequence, can significantly enhance the efficiency of manually constructing SynComs, while substantially reducing the error rate. This provides researchers in related fields with a convenient and practical experimental strategy.
Principles and Methods
SynComs are artificially constructed simple microbial consortia, typically containing between 3 to 7 species. To exhaustively construct all possible species combinations of n species, it is necessary to consider scenarios including zero species, one species, two species, multiple species, and up to n species. Taking n=10 as an example, the total number of species combinations N can be calculated using the following formula (Equation 1):

As shown in Figure 1, within this exhaustive construction scheme, the number of combinations without any species (blank control) is 1, single-species combinations are 10, two-species combinations are 45, three-species combinations are 120, and so on, totaling 1,024 combinations. Although blank control and single-species combinations may not be considered microbial communities in the traditional sense, they are essential controls for research and thus are important members of this SynCom system.
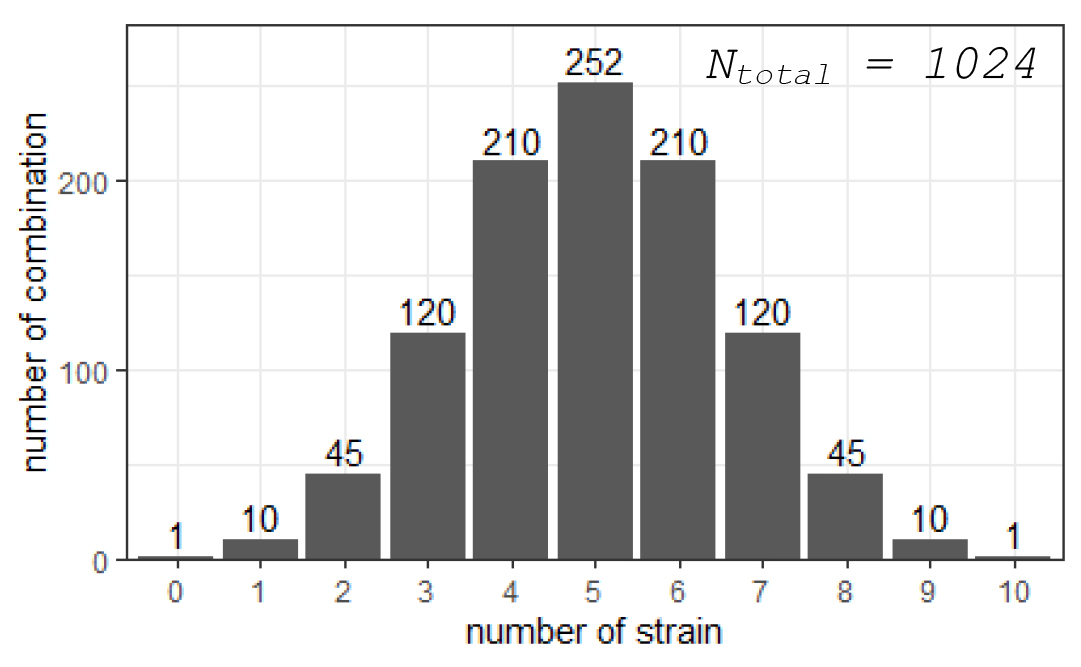
Figure 1. The number of SynCom combinations with different numbers of species when exhaustively constructing SynComs with 10 different strains.
More generally, if the total number of possible SynCom combinations constructed from n strains is denoted as N, then according to the binomial theorem, the value of N is equal to 2n (Equation 2). Therefore, the number of combinations of SynComs increases exponentially with the number of strains. Given this, while it may be relatively easy to manually construct dozens of different SynComs in a short time, the task becomes prone to errors without proper design when dealing with hundreds of different combinations as the number of species increases.

From a combinatorial mathematics perspective, 2n represents the total number of ways to select subsets from n elements. Each subset corresponds to a 0–1 vector, where 0 indicates the absence of an element and 1 indicates its presence. Thus, when these vectors are sorted, they can be orderly arranged on a 2(n-1) × 2(n-1) layout on a two-dimensional plane, providing important guidance for planning the order of additions during the construction of SynComs. Since there are no 2(n-1) × 2(n-1) microtiter plates in laboratories, and common microtiter plates are available in formats like 24-well, 96-well, and 384-well, experimental operations are orderly carried out on the available space of 24-well, 96-well, and 384-well plates by rows and columns. Each SynCom is given a unique id based on its position on the plate. Ultimately, this method can construct over a thousand different combinations of SynComs efficiently. When constructing SynComs on the latter two types of microtiter plates, using multi-channel pipettors can significantly improve work efficiency. To clearly identify the composition of the synthetic microbial communities in each well after addition, we also developed an R package ‘syncons’. This package can be run locally or accessed through cloud services, and in addition to providing SynComs ids, it also offers functions such as sampling process inquiry and data collection form generation.
Overall, this method aims to improve the efficiency of manually constructing SynComs using common laboratory materials and equipment, with careful planning of the addition order, while also reducing errors and avoiding cross-contamination, providing researchers in related fields with a convenient and practical experimental strategy.
Materials and reagents
24-well plate (Corning, catalog number: 3524)
96-well plate (Corning, catalog number: 3599)
384-well plate (Axygen®, catalog number: P-384-240SQ-C-S)
Pipette tip (Biosharp, catalog number: BS-200-T)
Reagent Reservoir (Biosharp, catalog number: BS-PS50-RR)
Culture media (TSB media, LB media, etc.) (BD, catalog number: GD-211825)
Strains
Different strains are distinguished by their IDs, herein represented as Strain S1 through S11, totaling eleven distinct bacterial strains.
Equipment
Shaking incubator (Huamei Biotech, catalog number: THZ-701B)
Single-channel pipette (Thermo ScientificTM, catalog number: 4651140N)
8-channel pipette (Thermo ScientificTM, catalog number: 4661010N)
16-channel pipette (EVOLVE, catalog number: 3044)
Laminar Flow Cabinet (AIRTECH, catalog number: ES-A01-VS-1300L-U)
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
