Advanced Search
Utility of molecular characterization in the diagnosis of chronic and recurrent dermatophytosis.
Published: Aug 19, 2022 DOI: 10.21769/p1871 Views: 618
UTILITY OF MOLECULAR CHARACTERIZATION IN THE DIAGNOSIS OF CHRONIC AND RECURRENT DERMATOPHYTOSIS
Aditi. P. Warghade 1, Dr. Gargi Mudey2
Department of Microbiology, Jawaharlal Nehru Medical College, Datta Meghe Institute of Medical Sciences, Sawangi(Meghe), Wardha,India.1
Department of Microbiology, Jawaharlal Nehru Medical College, Datta Meghe Institute of Medical Sciences, Sawangi(Meghe), Wardha,India2
Abstract
Dermatophytes are the keratinophilic fungi which infect humans and is the most recurring type of disease. The high level of transmissibility creates an epidemiological risk and emphasises the significance of these illnesses. However, a growing number of reports describing dermatophytes which can cause deep infections in Diabetic& Immunocompromised Patients, as well as in those with Immunodeficiency, by invading deep layers like the dermis and hypodermis. Despite the prevalence and significance of dermatophytes in clinical mycology, it is not always possible to accurately diagnose this specific infection due to its overlapping structures among species of dermatophytes.Since it is difficult to identify species that exhibit weak characteristics in the morphological highlights, identification of the dermatophyte is often relied on its morphological analysis, which is a laborious process and demands skill. The massive shift in genetic variation, the source of infection, and epidemiological research can be discovered using molecular approaches. Therefore, the development of an accurate laboratory test for dermatophyte species identification is essential for the prevention and efficient management of dermatophytoses. One such methodology allows use of PCR technology which has many methods for molecular level characterization which is rapid, efficient, and capable of producing DNA polymorphisms specific to various dermatophyte species based on distinctive band patterns seen by agarose gel electrophoresis. The RAPD-PCR approach will be used in this study protocol to molecularly characterise the dermatophytes for precise speciation of the sample. In addition to improving knowledge of fungal biology and pathology with a focus on adaptive mechanisms to combat difficult conditions from host counteractions, there is a need to improve awareness of the importance of these diseases through accurate epidemiological data. The advantages of molecular approaches for characterising objects over traditional methods are their sensitivity and specificity.
Keywords: Dermatophytosis, Molecular Characterisation, RAPD-PCR,Electrophoresis.
Background:
Dermatophytes are responsible for different clinical manifestations ranging from superficial as tinea corporis, tinea capitis, tinea pedis, and tinea unguium to subcutaneous affection ( Diongue K et al.,2019). These diseases can range in severity clinically from moderate to severe depending on the host's immune system, the virulence of the strain, and other environmental factors (Weitzman I et al.,1995; Havlickova B et al .,2005; Gnat S et al .,2018). Although dermatophytoses harm people throughout, they are more common in tropical regions due to high temperatures and humidity (Taplin et al.,2020) .Age, sex, the time of year, socioeconomic and cultural circumstances, and geographic location are all factors that might influence the development of dermatophytosis (Iorio R et al.,2007) .Potassium hydroxide (KOH) direct microscopy followed by selective medium culture is the standard procedure for dermatophyte screening in clinical samples. A quick screening technique for fungal structures is microscopy performed directly on clinical specimens, but this method lacks specificity( Garg J et al.,2013). Dermatophyte isolates can be identified to genus/species by phenotypic methods, based on colonial appearance, microscopic examinations, and biochemical tests such as growth patterns(Aggarwal R et al.,2020). Morphological identification of dermatophyte species in cultures is sometimes difficult or uncertain because there are variations from one isolate to another and overlapping characters between species(Verrier J et al.,2017). Molecular-based methods rely on identifying genotypic variations in pathogenic organisms( Bergman A et al., 2013;Kondori N et al 2010;Paugam A et al.,2013). Molecular methods are being used to identify dermatophytes since they are more precise than conventional techniques (Tartor YH et al., 2019).The development of molecular diagnostics were encouraged by the conventional techniques for identifying dermatophytes which are imprecise and have a delayed diagnostic character.Techniques that permit for both the early and accurate detection of dermatophytosis in order to provide timely antifungal treatment that prevents generic over-the-counter medication (Begum J et al., 2020) Therefore, it is essential to create more reliable dermatophyte identification techniques( Gräser Y et al.,2007). The current study's objective is to utilize RAPD-PCR method to identify and distinguish between the strains of fungi present in clinical isolates that cause chronic recurrent dermatophytosis.
Materials and Reagents
Culture media Sabouraud Dextrose Agar (SDA) (HiMedia, 2021-2022,M063).
Culture media Sabouraud Chloramphenicol + Cycloheximide Agar (HiMedia, 2021-2022,M664)
Culture media Dermatophyte Agar Base (HiMedia, 2021-2022,M188)
Dermato Supplement (HiMedia, 2021-2022, PD015)
Culture Media Corn meal agar (HiMedia, 2021-2022,M146)
Distilled Water
Potassium Hydroxide Pellets (HiMedia, 2021-2022, MB262)
Normal Saline
10% Glycerol (HiMedia, 2021-2022, MB060)
Lactophenol Cotton Blue Stain (HiMedia, 2021-2022, GRM8537)
Polyvinyl Alcohol (HiMedia, 2021-2022, GRM6170)
Ethyl Alcohol
Sodium Hypochlorite Solution (HiMedia, 2021-2022, AS102)
dNTP mix,40mM (Ready to use mix of dATP, dCTP, dGTP, dTTP solutions)(HiMedia,2021-2022,MBT187-1ml)
Hi-Temp PCR Master mix (HiMedia,2021-2022, MBT119-100R)
Primers (HiMedia)
AllPrep® Fungal DNA isolation kit (QIAGEN,Catalogue no-47154)
PCR Block Plates (Himedia,2021-2022)
Pipette
Pipette tips (10µl,20µl,100µl,1000µl)
Microscopic glass slides
Cover slip
Inoculating loop
Teasing needle
Petri dishes
Forceps
Glass Beakers
Flasks
Glass rod
Bunsen Burner
Gel electrophoresis chamber
Equipment
Light Microscope
Autoclave
PCR
Biosafety Cabinet
BOD Incubator
Electronic Weighing Machine
Procedure
A. Sample collection:
1. Clean the area of sample collection with 70% alcohol.
2. For Skin samples,scrape the lesion around the corners using scalpel blade or glass slide
3. For Nail samples, collect the nail clippings using nail cutter.
4. For hair samples, Pluck the hair from shaft having lesion.
B. Sample Processing:
1. For skin samples dissolve the sample in 10% KOH and for Nail clipping dissolve in 40% KOH for microscopic observation.
2. Inoculate the sample on Dermatophyte Test Medium as well as Sabouraud's dextrose agar containing Chloramphenicol&Cycloheximide.
3. Incubate the samples in BOD incubator, observe after growth.
4. Lactophenol cotton Blue staining and Slide culture technique are used to view morphology and colony characteristics of the sample after growth.
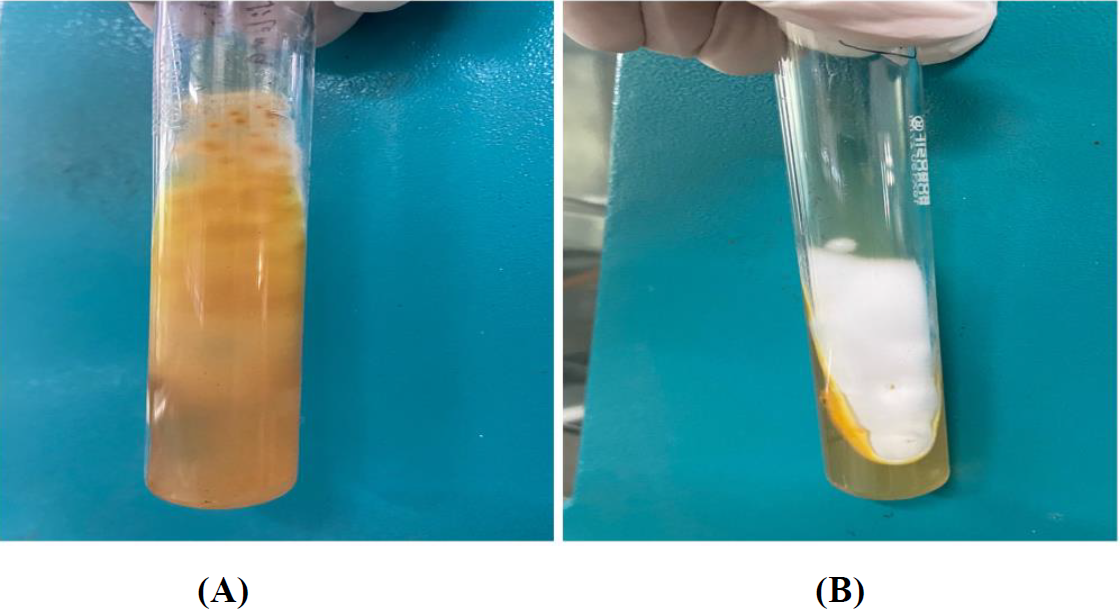
Figure A: - ATCC strain of Trichophyton Interdigitale on Reverse Side of SDA
Figure B: - ATCC strain of Trichophyton Interdigitale on Front side of SDA
C. Application of RAPD-PCR method for molecular characterization:
1. Standardise molecular assay using the standard strains of Trichophyton (D15P127, CBS 118892, UCMS-IGIB-CI11), Microsporum (ATCC 36299) and laboratory confirmed strains.
2. AllPrep® Fungal DNA isolation kit is used for DNA isolation from fungal cultures.
3. Primer required for the RAPD-PCR reaction is synthesized by Beacon designer probe/primer designer software from GeneX India Biosciences Velachery, Chennai, Tamil Nadu.
4. PCR assay mixture, reaction buffer, dNTP's, each primer set (GACA4) and novel primer (CTGT3), DNA template, using these PCR reaction cycles are carried out.
a) 39 cycles
b) Denaturation at 93° - 1-minute,
c) Annealing step at 50° -1-minute,
d) -Extension step at 72°- 1 minute
e) Finalextension step at 72°- 7 min
5. Final PCR products are separated in 0.5X (Tris Borate-Ethylene diamine tetra acetic acid)Buffer&1% Agarose and stained with the Ethidium-Bromide solution & then the image are obtained.
6. Interspecies and intraspecies Pattern and polymorphism for known strains studied.
Recipes
1. Sabouraud Chloramphenicol + Cycloheximide Agar- (For 1000ml)
a) 10gms of Peptone
b) 20gms of Dextrose (Glucose)
c) 50mg Chloramphenicol
d) 500mg Cycloheximide
e) 15gms Agar
f) Final pH will be adjusted to 6.8±0.2 (at 25 °C)
g) By autoclaving the Media for 15 minutes at 121°C and 15 lbs of pressure, sterilise. Mix thoroughly, then transfer it to sterilised Petri dishes.
h) https://himedialabs.com/TD/M664.pdf
2. Agar Base D.T.M. (Dermatophyte Test Medium) (For 500ml)
a) 10gms Soya peptone
b) 10gms Dextrose (Glucose)
c) 0.200 gms Phenol red
d) 20.00gms Agar
e) Final pH will be adjusted to 5.5±0.2 ( at 25°C)
f) In 500 ml of distilled or purified water, suspend 20.10 grams. To completely dissolve the medium, heat it until it boils. Autoclave for 15 minutes at 121°C & 15 lbs of pressure, sterilise. to 45–50 °C. Add the rehydrated contents of one Dermato Supplement vial aseptically (FD015). Before putting into sterilised Petri plates, mix thoroughly.
https://himedialabs.com/TD/M188.pdf
3. Corn Meal Agar
a) 50.00 gms Corn meal infusion form
b) 15.00gms Agar
c) Final pH will be adjusted to 6.0±0.2 ( at 25°C)
d) In 1000 litres of distilled or purified water, dissolve 17 grams. To completely dissolve the medium, heat it until it boils. Add 1% polysorbate 80 if desired. Autoclave for 15 minutes at 121°C & 15 lbs of pressure, sterilise. to 45–50 °C. Mix thoroughly, then transfer to sterile Petri dishes.
e) https://himedialabs.com/TD/M146.pdf
Ethics
The following doctoral research project has been approved by the institutional ethics committee to be carried out at Acharya Vinoba Bhave Rural Hospital Sawangi (Meghe) Wardha and Jawaharlal Nehru Medical College.
References
- Diongue K, Boye A, Bréchard L, Diallo MA, Dione H, Ndoye NW, Diallo M, Ranque S, Ndiaye D. Dermatophytic mycetoma of the scalp due to an atypical strain of Microsporum audouinii identified by MALDI-TOF MS and ITS sequencing. Journal de Mycologie Médicale. 2019 Jun 1;29(2):185-8.
- Weitzman I, Summerbell RC. The dermatophytes. Clinical microbiology reviews. 1995 Apr;8(2):240-59.
- Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008 Sep;51:2-15.
- Gnat S, Łagowski D, Nowakiewicz A, Trościańczyk A, Zięba P. Infection of Trichophyton verrucosum in cattle breeders, Poland: A 40‐year retrospective study on the genomic variability of strains. Mycoses. 2018 Sep;61(9):681-90.
- Taplin, D. Dermatophytosis in Vietnam. Cutis 2001, 67, 19–20.
- Iorio R, Cafarchia C, Capelli G, Fasciocco D, Otranto D, Giangaspero A. Dermatophytoses in cats and humans in central Italy: epidemiological aspects. Mycoses. 2007 Nov;50(6):491-5.
- Garg J, Tilak R, Singh S, Gulati AK, Garg A, Prakash P, Nath G. Evaluation of pan-dermatophyte nested PCR in diagnosis of onychomycosis. Journal of clinical microbiology. 2007 Oct;45(10):3443-5
- Aggarwal R, Targhotra M, Sahoo PK, Chauhan MK. Onychomycosis: Novel strategies for treatment. Journal of Drug Delivery Science and Technology. 2020 Jun 1;57:101774.
- Verrier J, Monod M. Diagnosis of dermatophytosis using molecular biology. Mycopathologia. 2017 Feb;182(1):193-202.
- Bergman A, Heimer D, Kondori N, Enroth H. Fast and specific dermatophyte detection by automated DNA extraction and real-time PCR. Clinical Microbiology and Infection. 2013 Apr 1;19(4):E205-11.
- Kondori N, Abrahamsson AL, Ataollahy N, Wennerås C. Comparison of a new commercial test, dermatophyte‑PCR kit, with conventional methods for rapid detection and identification of Trichophyton rubrum in nail specimens. Med Mycol 2010;48:1005‑8.
- Paugam A, L’ollivier C, Viguié C, Anaya L, Mary C, de Ponfilly G, et al. Comparison of real‑time PCR with conventional methods to detect dermatophytes in samples from patients with suspected dermatophytosis. J Microbiol Methods 2013;95:218‑22
- Tartor YH, Abo Hashem ME, Enany S. Towards a rapid identification and a novel proteomic analysis for dermatophytes from human and animal dermatophytosis. Mycoses. 2019 Dec;62(12):1116-26.
- Begum J, Mir NA, Lingaraju MC, Buyamayum B, Dev K. Recent advances in the diagnosis of dermatophytosis. Journal of basic microbiology. 2020 Apr;60(4):293-303
- Gräser Y, Fröhlich J, Presber W, de Hoog S. Microsatellite markers reveal geographic population differentiation in Trichophyton rubrum. Journal of medical microbiology. 2007 Aug 1;56(8):1058-65.
- Shehata AS, Mukherjee PK, Aboulatta HN, El Akhras AI, Abbadi SH, Ghannoum MA. Single-step PCR using (GACA) 4 primer: utility for rapid identification of dermatophyte species and strains. Journal of clinical microbiology. 2008 Aug;46(8):2641-5.
- https://himedialabs.com/TD/M664.pdf
- https://himedialabs.com/TD/M188.pdf
- https://himedialabs.com/TD/M146.pdf
- Bergman A, Heimer D, Kondori N, Enroth H. Fast and specific dermatophyte detection by automated DNA extraction and real-time PCR. Clinical Microbiology and Infection. 2013 Apr 1;19(4):E205-11.
- Lund A, Bratberg AM, Næss B, Gudding R. Control of bovine ringworm by vaccination in Norway. Veterinary immunology and immunopathology. 2014 Mar 15;158(1-2):37-45.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
