Advanced Search
miRNA Stem-loop RT-qPCR Method
Last updated date: Nov 8, 2024 Views: 197 Forks: 0
1.Introduction to miRNA
Small RNAs are regulatory RNA molecules ranging from 19 to 28 nucleotides (nt) in length, mainly including micro RNAs (miRNAs) and short interfering RNAs (siRNAs). Among them, miRNAs have become one of the new research hotspots following siRNAs and were listed among the top ten scientific achievements of the year by the American journal Science in both 2002 and 2003.
miRNAs are part of longer RNA sequences and, like siRNAs, are relatively short single-stranded small molecular RNAs, generally originating from non-coding regions of the chromosomes. They are processed from precursors that are about 70 nt in size and can form hairpin structures. They inhibit the translation of their target mRNA molecules by binding to the 3' untranslated region (3' UTR) at the 3' end of the mRNA.
In the public miRNA database miRBase (http://www.mirbase.org/), there are already more than 10,000 miRNA sequences from various species.
2.Experimental Principle
Based on the spatial structure of the stem-loop primer to enhance the specificity of reverse transcription, thereby reverse transcribing specific small RNAs. In the real-time quantitative process, the stem-loop structure is opened, and quantification is performed using SYBR dye with universal and specific primers.
Experimental Objective: To detect the expression levels of small RNAs in various plant tissues, especially in materials with limited samples.
Keywords: Small RNA, Stem-loop RT, Real-time PCR, Stem-loop structure, SYBR
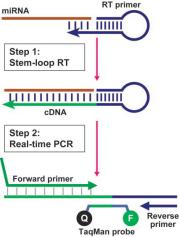
3.Materials and Reagents
Various sizes of pipette tips (RNase free and general tips)
96-well plates
DEPC water
DNase I (NEB, catalog number: M0303S)
SuperScript® III Reverse Transcriptase (Invitrogen, catalog number: 18080-044)
RNase Inhibitor (Takara, catalog number: D2313A)
SYBR Premix Ex TaqTM (Takara, catalog number: DRR041A)
EDTA
Instruments and Equipment:
Pipettes
9700 PCR System (ABI)
7500 Real-Time PCR System (ABI)
Centrifuge
4.Experimental Procedure
4.1Plant RNA Extraction (Kit Method, e.g., using RNAsimple Total RNA Extraction Kit, DP419)
4.1.1 Take a high-temperature, high-pressure disinfected steel ball and place it in an RNase-free 2ml centrifuge tube. Add approximately 0.2g of the plant tissue under study. Then add 1 ml of Buffer RZ and homogenize the sample using a homogenizer.
4.1.2 Place the homogenized sample at room temperature for 5 minutes to allow complete separation of nucleic acid-protein complexes.
Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 5 minutes, collect the supernatant, and transfer it to a new RNase-free centrifuge tube.
4.1.3 Add 200 μl of chloroform, close the tube cap tightly, and vortex vigorously for 15 seconds. Let it sit at room temperature for 3 minutes.
4.1.4 Centrifuge at 4°C, 12,000 rpm (~13,400×g) for 10 minutes. The sample will separate into three layers: a yellow organic phase, a middle layer, and a colorless aqueous phase. RNA is mainly in the aqueous phase, which is approximately 50% of the volume of the Buffer RZ reagent used. Transfer the aqueous phase to a new tube for the next step.
4.1.5 Slowly add 0.5 times the volume of anhydrous ethanol and mix well (a precipitate may form at this point). Transfer the solution and precipitate together into the RNase-Free Columns CR3 set, centrifuge at 4°C, 12,000 rpm (~13,400×g) for 30 seconds. If you cannot transfer all the solution and mixture into the RNase-Free Columns CR3 set at once, please do it in two steps, centrifuging at 4°C, 12,000 rpm (~13,400×g) for 30 seconds each time, discarding the waste in the collection tube.
4.1.6 Add 500μl of protein removal liquid RD to the RNase-Free Columns CR3 set (please check if ethanol has been added before use), centrifuge at 4°C, 12,000 rpm (~13,400×g) for 30 seconds, discard the waste, and place the RNase-Free Columns CR3 set into the collection tube.
4.1.7 Add 500 μl of wash liquid RW to the adsorption column CR3 (please check if ethanol has been added before use), let it stand at room temperature for 2 minutes, centrifuge at 4°C, 12,000 rpm (~13,400×g) for 30 seconds, and discard the waste.
4.1.8 Repeat step 4.1.7.
4.1.9 Place the adsorption column into a 2 ml collection tube, centrifuge at 4°C, 12,000 rpm (~13,400×g) for 2 minutes to remove residual liquid.
Note: The purpose of this step is to remove residual wash liquid from the adsorption column. After centrifugation, place the adsorption column at room temperature for a while, or on a super clean bench for ventilation, to dry thoroughly.
4.1.10 Transfer the adsorption column CR3 into a new 1.5 ml centrifuge tube, add 30-100 μl of RNase-Free ddH2O, let it stand at room temperature for 2 minutes, and centrifuge at 4°C, 12,000 rpm (~13,400×g) for 2 minutes.
4.1.11 Repeat step 4.1.10, then measure the concentration (OD260/280 value should be between 1.8-2.0), and store at -80°C.
4.2.The design method for the stem-loop primer and real-time PCR primer for miRNA
The design method for the stem-loop primer and real-time PCR primer for miR1440b is as follows:
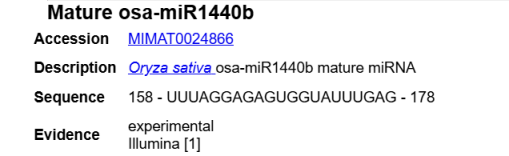
Universal primer (forming a stem-loop): GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC (the red part is complementary, and the black part forms the stem-loop).
osa-miR1440b stem-loop RT prime: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTCAAA
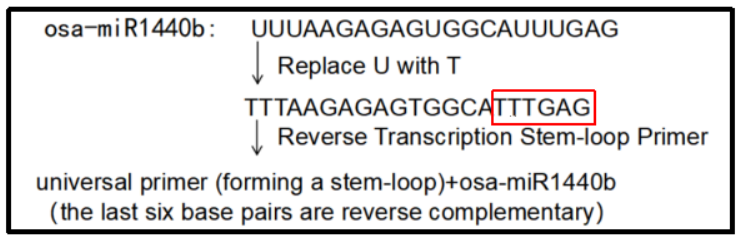
Universal primer R(a segment from the stem-loop universal primer): GTGCAGGGTCCGAGGT
Forward prime(The remaining part of the miRNA sequence after removing the 3' terminal 6 bases is used as the upstream primer, and C/G is added to the 5' end to complement the TM to 60 ℃): CCCGGTTTAAGAGAGTGGCA
(Designs can also be made using websites:https://www.sangon.com/newPrimerDesign)
4.3.Stem-loop reverse transcription reaction(Sangon Biotech,B532453 )
In a 200 µl centrifuge tube, add the following reaction mixture:
| 2×miRNA L-RT Solution mix | 10 µl |
| miRNA L-RT Enzyme mix | 1.5 µl |
| micro RNA | 3-4 µg/200 ng |
| Stem-loop primer(10 µM) | 1 µl |
Gently mix and centrifuge for 3-5 seconds. Incubate the reaction mixture at 16°C for 30 minutes, then at 37°C for 30 minutes, and finally at 85°C for 5 minutes to inactivate the enzyme, then store at 4°C.
4.4.miRNA Fluorescence Quantitative PCR(Sangon Biotech,B532461 )
In a 200 µl centrifuge tube, add the following reaction mixture:
| 2×miRNA qPCR master mix | 10 µl |
| Forward PrimerM ,10 µM | 0.5 µl |
| Reverse Primer,10 µM | 0.5 µl |
| Template DNA | 1-2 μl |
| OX Reference Dye(L)/(H) | 1 μl |
| RNase-free water | up to 20 μl |
Gently mix and centrifuge the PCR reaction mixture for 3-5 seconds. Begin with an initial denaturation at 95°C for 3-5 minutes, then perform the two-step PCR cycling with denaturation at 95°C for 10-15 seconds and annealing/extension at 60-68°C for 20-40 seconds, repeating these steps for 25-35 cycles. Finally, conduct a final extension at 72°C for 5-10 minutes, and then inactivate the enzyme by cooling the reaction to 4°C for storage or further analysis.
Attention:
Typically, a primer concentration of 0.25 μM can yield good results, and a final concentration range of 0.1-1.0 μM can be used as a reference for setting. If the amplification efficiency is not high, the primer concentration can be increased; if primer dimers are present, the primer concentration can be decreased to optimize the reaction system.
Gradient dilution of the template can be performed to determine the optimal amount of template to use.
ROX Reference Dye should be selected based on the model of the instrument used:
ROX Reference Dye(H) is suitable for the following models:
ABI Prism7000/7300/7700/7900, Eppendorf Realplex 4, ABI Step One, ABI Step One Plus.
ROX Reference Dye(L) is suitable for the following models:
ABI Prism7500, ABI Prism7500 Fast, Stratagene Mx3000/Mx3005P, Corbett Rotor Gene 3000.
The following models do not require ROX correction:
Roche LightCycler480, Roche Light Cycler96, MJ Research Chromo4, Opticon (II), Bio-Rad iCycler iQ, iQ5, Bio-Rad CFX96, Corbett Rotor Gene 6000, Eppendorf Realplex 2.
4.5.qPCR Data Calculation of Relative Gene Expression (-ΔΔCt Method)
When calculating relative gene expression levels from qPCR data, the -ΔΔCt method is a commonly used approach. Here is the specific calculation process:
Calculate ΔCt: For each sample, the ΔCt value is obtained by subtracting the Ct value of the reference gene (usually a housekeeping gene, such as actin) from the Ct value of the target gene. The formula is:
![]()
Calculate ΔΔCt: The ΔΔCt value is the difference between the ΔCt value of the experimental group and the average ΔCt value of the control group (or baseline sample). The formula is:
![]()
Calculate 2^(-ΔΔCt): Finally, substitute the ΔΔCt value into the formula to calculate 2^(-ΔΔCt), which gives the relative expression level. The formula is:
![]()
Example calculation process:
Suppose we have three samples, a control group (Control) and two experimental groups (Sample1 and Sample2), with the target gene being 595 and the reference gene being actin. The Ct values are as follows:
Control: 595 Ct = 25, actin Ct = 20
Sample1: 595 Ct = 28, actin Ct = 21
Sample2: 595 Ct = 30, actin Ct = 22
First, calculate ΔCt:
ΔCt(Control) = 25 - 20 = 5
ΔCt(Sample1) = 28 - 21 = 7
ΔCt(Sample2) = 30 - 22 = 8
Next, calculate ΔΔCt:
ΔΔCt(Sample1) = 7 - 5 = 2
ΔΔCt(Sample2) = 8 - 5 = 3
Finally, calculate 2^(-ΔΔCt):
Relative Expression(Sample1) = 2^(-2) ≈ 0.25
Relative Expression(Sample2) = 2^(-3) ≈ 0.125
This means that the expression level of the target gene in Sample1 is 0.25 times that of the control group, and in Sample2, it is 0.125 times that of the control group. Using this method, we can compare the relative expression levels of the target gene in different samples.
References:
Chen, C., Ridzon, D. A., Broomer, A. J., Zhou, Z., Lee, D. H., Nguyen, J. T., Barbisin, M., Xu, N. L., Mahuvakar, V. R., Andersen, M. R., Lao, K. Q., Livak, K. J., and Guegler, K. J. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20): e179.
Shen, J. Q., Li, Y. and Xiong, L. Z. (2018). Stem-loop RT real-time PCR. Bio-101 e1010118.
Doi: 10.21769/BioProtoc.1010118. (in Chinese)
Livak, K.J. & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25(4):402–408. doi: 10.1006/meth.2001.1262 (2001).
Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45 (2001).
- Wang, S(2024). miRNA Stem-loop RT-qPCR Method. Bio-protocol Preprint. bio-protocol.org/prep2747.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
