- EN - English
- CN - 中文
Detection of DNA Methylation Changes Surrounding Transposable Elements
转座子侧翼区域DNA甲基化水平的检测
发布: 2013年06月05日第3卷第11期 DOI: 10.21769/BioProtoc.784 浏览次数: 11687
评审: Tie Liu

相关实验方案

使用Brick into the Gateway (BiG) 实验方法快速克隆细菌基因
Flaviani G. Pierdoná [...] Fabio T. S. Nogueira
2022年12月20日 2334 阅读
Abstract
Transposable elements (TEs) are a major component of all genomes, thus the epigenetic mechanisms controlling their activity is an important field of study. Cytosine methylation is one of the factors regulating the transcription and transposition of TEs, alongside Histone modifications and small RNAs. Adapter PCR-based methods [such as Amplified Fragment Length Polymorphism (AFLP)] have been successfully used as high-throughput methods to genotype un-sequenced genomes. Here we use methylation-sensitive restriction enzymes, in combination with PCR on adaptor-ligated restriction fragments, to evaluate epigenetic changes in TEs between genomic DNA samples.
Keywords: DNA methylation (DNA甲基化)Materials and Reagents
- Two oligonucleotides which form the double-stranded adapter, with an overhang complementary to the overhang of the restriction enzyme used. In the case of HpaII or MspI, the overhang is a 5' CG, and the adapter sequences are 5'-GATCATGAGTCCTGCT-3' and 5'-CGAGCAGGACTCATGA-3'. The two nucleotides at the 5' end of the latter oligonucleotide will constitute the 5' CG overhang, after hybridization of the two sequences (black rectangles in Figure 1, Shaked et al., 2001). These oligonucleotides should be designed such that they do not resemble know sequences in the examined species.
- Pre-selective primers, one complementary to the adapter with the addition of a G nucleotide at the 3' end (5'-ATCATGAGTCCTGCTCGG-3'; primer P2 in Figure 1), and the other complementary to the TE of interest (primer P1 in Figure 1). The TE-specific primer should be designed as a reverse-complement of the 5' end of the TE with a Tm=60 °C, between 30-50 bp into the TE (to allow for sequence validation in downstream assays). Restriction enzyme recognition sites (CCGG) between the primer and the 5' end of the TE should be avoided.
- Selective primers, one identical to the above TE-specific primer with the addition of a fluorescent tag (e.g. 6-FAM) or radioactive tag (end label with 32P), and one similar to the pre-selective primer complementary to the adapter with the addition of random nucleotides at the 3' end (e.g. 5'-CATGAGTCCTGCTCGGTCAG-3', includes an extra TCAG at the 3' end).
- NaCl
- T4 DNA ligase and buffer (New England Biolabs, catalog number: M0202 )
- Restriction enzymes HpaII and MspI (New England Biolabs, catalog number: R0171 and R0106 )
- Taq DNA polymerase and Taq DNA polymerase buffer (EURx, catalog number: E2500 )
- MgCl2
- dNTP mix
- Polynucleotide Kinase (PNK) enzyme and PNK buffer (New England Biolabs, catalog number: M0201 )
- Gamma-phosphate (32P)-labeled ATP (or fluorescently-labeled primers)
- GS-500 ROX-labeled size standard (for fluorescently-labeled products only) (Applied Biosystems)
- Hi-Di Formamide (for fluorescently-labeled products only) (Applied Biosystems, catalog number: 4311320 )
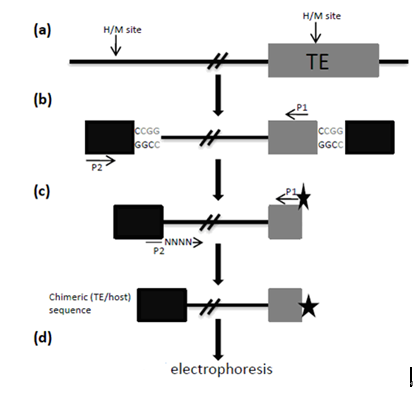
Figure 1. An overview of the TMD method, adapted from (Yaakov and Kashkush, 2011). The method steps include: (a) Restriction of genomic DNA with HpaII (H) or MspI (M); (b) The first round of PCR amplification, using a primer from within the TE (P1) and a primer from the adapter (P2); (c) The second round of PCR amplification, using primer P1 from within the TE, labeled with a radioactive or fluorescent tag, and primer P2 with the addition of random nucleotides at the 3' end; and (d) Electrophoresis of the resulting PCR amplicons on a polyacrylamide gel (in the case of a radioactive tag) or in a capillary fluorescence detection machine (in the case of a fluorescent tag).
Equipment
- Thermal cycler
- Agarose gel electrophoresis machine
- 43 cm PAGE machine (Thermo Scientific Owl Aluminum-Backed Sequencer S3S, for radiolabeled products only)
- Capillary electrophoresis machine (such as the Applied Biosystems 3730xl DNA analyzer; for fluorescently-labeled products only)
Procedure
文章信息
版权信息
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Yaakov, B. and Kashkush, K. (2013). Detection of DNA Methylation Changes Surrounding Transposable Elements. Bio-protocol 3(11): e784. DOI: 10.21769/BioProtoc.784.
分类
系统生物学 > 表观基因组学 > DNA 甲基化
植物科学 > 植物分子生物学 > DNA > DNA 修饰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link