- EN - English
- CN - 中文
A Rapid and Cost-Effective Pipeline to Identify and Capture BGCs From Bacterial Draft Genomes
一种快速且经济的流程,用于从细菌草图基因组中识别并捕获生物合成基因簇
发布: 2025年12月20日第15卷第24期 DOI: 10.21769/BioProtoc.5549 浏览次数: 778
评审: Alba BlesaAnonymous reviewer(s)

相关实验方案

使用Brick into the Gateway (BiG) 实验方法快速克隆细菌基因
Flaviani G. Pierdoná [...] Fabio T. S. Nogueira
2022年12月20日 2364 阅读

番茄内生菌Streptomyces sp. VITGV100中吲哚类代谢产物的合成与提取分析
Veilumuthu Pattapulavar [...] John Godwin Christopher
2025年07月20日 2424 阅读
Abstract
The exploration of microbial genomes through next-generation sequencing (NGS) and genome mining has transformed the discovery of natural products, revealing an immense reservoir of previously untapped chemical diversity. Bacteria remain a prolific source of specialized metabolites with potential applications in medicine and biotechnology. Here, we present a protocol to access novel biosynthetic gene clusters (BGCs) that encode natural products from soil bacteria. The protocol uses a combination of Oxford Nanopore Technology (ONT) sequencing, de novo genome assembly, antiSMASH for BGC identification, and transformation-associated recombination (TAR) for cloning the BGCs. We used this protocol to allow the detection of large BGCs at a relatively fast and low-cost DNA sequencing. The protocol can be applied to diverse bacteria, provided that sufficient high-molecular-weight DNA can be obtained for long-read sequencing. Moreover, this protocol enables subsequent cloning of uncharacterized BGCs into a genome engineering-ready vector, illustrating the capabilities of this powerful and cost-effective strategy.
Key features
• This protocol enables bioprospection through cloning of a novel BGC identified in an ONT bacterial draft genome.
• A combination of ONT sequencing, antiSMASH, and TAR cloning can be used to clone BGCs from bacteria into a vector.
• Cost-effective strategy for the discovery of BGCs of diverse natural product classes, including nonribosomal peptides, polyketides, and RiPPs.
• Overnight sequencing in-house using cheap and easy-to-use instruments such as MinION, which allows multiplexing.
Keywords: Biosynthetic gene clusters (生物合成基因簇)Graphical overview
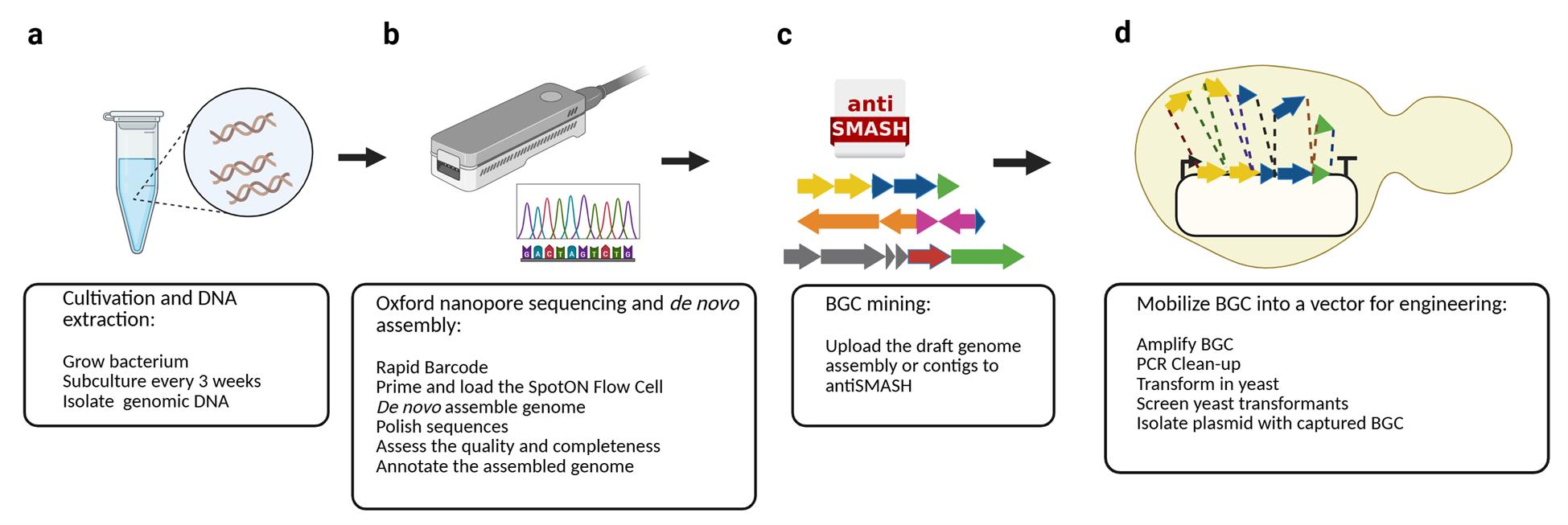
Overview of the pipeline. The protocol consists of four main steps: (a) Cultivation and isolation of genomic DNA of the bacterium of interest, (b) Oxford Nanopore sequencing and de novo genome assembly, (c) BGC mining using antiSMASH, and (d) biosynthetic gene cluster (BGC) mobilization into a genome engineering-ready vector via PCR amplification and TAR cloning in yeast.
Background
Microbial natural products are chemically diverse secondary metabolites with ecological functions and major applications in drug discovery and biotechnology. Many of these compounds are produced by soil and marine bacteria, yet their exploitation is often hampered by difficulties in cultivation and expression under laboratory conditions [1]. A key advance in natural product research has been the identification of biosynthetic gene clusters (BGCs), which encode the enzymatic machinery for metabolite production, regulation, transport, and, if necessary, resistance to the BGC metabolite. Genome sequencing and mining have accelerated the discovery of BGCs that encode enzymes for the production of secondary metabolites. However, there is still a limited number of genomes available from bacteria rich in secondary metabolites, such as myxo- and actinobacteria [2] in comparison to bacterial genome sequences of human pathogens.
The discovery of novel natural products increasingly relies on the systematic mining of BGCs. While many bacteria, such as myxobacteria and actinobacteria, are renowned for their richness in biosynthetic potential, their limited amenability to genetic manipulation makes direct engineering a major challenge. As an alternative, heterologous expression of BGCs in well-characterized laboratory microorganisms has emerged as a promising strategy [3–7]. However, this approach involves overcoming several technical bottlenecks: from sequencing and identifying large, GC-rich, and repetitive genomes, to cloning and refactoring BGCs, introducing them into suitable hosts, achieving heterologous production, and finally isolating and characterizing the resulting compounds. Recent technological advances have addressed many of these hurdles. For instance, genome mining tools such as antiSMASH have streamlined BGC identification [8], while CRISPR-based approaches have allowed the refactoring of large gene clusters [9,10]. Similarly, versatile genome engineering platforms such as chassis-independent recombinase-assisted genome engineering (CRAGE) facilitate transfer into diverse bacterial hosts [11]. Current heterologous expression strategies vary in their reliance on genome sequence quality: while de novo DNA synthesis requires highly accurate BGC sequences [12–14], methods such as transformation-associated recombination (TAR) cloning [15,16] and Cas9-assisted targeting of chromosome segments (CATCH) [17] are compatible with draft genomes, depending on oligonucleotides for primer or gRNA design. Draft genome sequencing itself has become increasingly accessible with ONT long-read platforms due to reduced costs and multiplexing capability. Generating highly accurate assemblies of large, repetitive, GC-rich genomes still requires combined short- and long-read approaches, which remain costly when scaled to numerous strains. However, draft genome sequencing allows the detection and cloning of a bacterial BGC [18]. Several technologies for whole genome sequencing exist, including Illumina, PacBio, and ONT. While PacBio sequencing offers higher accuracy, ONT provides a more affordable and scalable option, delivering data of sufficient quality for BGC cloning, as shown in Campos-Magaña et al. [18]. Alternatively, hybrid ONT/Illumina strategies can be employed to generate high-quality genome assemblies [19]. However, currently, the cost of a hybrid approach is more expensive than sequencing only using ONT. Furthermore, ONT sequencing prices can be even lowered by performing it in-house using cheap and easy-to-use instruments such as MinION, which allows multiplexing. This protocol enables the generation of a draft genome using ONT sequencing and extracted bacterial genomic DNA. This protocol combines ONT sequencing with antiSMASH and TAR cloning for scalable discovery and capture of BGCs. We used TAR cloning in yeast due to its ability to clone large constructs via in vivo homologous recombination, allowing us to precisely clone BGCs identified in Oxford Nanopore draft genomes, despite the inherent limitations and reduced accuracy of this sequencing platform. This strategy is also compatible with subsequent refactoring of captured BGCs, an important step that involves replacing native regulatory elements with more suitable ones for heterologous expression, since the final vector with the captured BGC can serve as input DNA for multiplex editing using CRISPR-Cas9 [20–22] and mobilization into diverse heterologous hosts using CRAGE [11]. Such a streamlined, rapid, and low-cost approach applied to the large collections of non-sequenced bacteria will enable exploration of BGCs in microbes.
Materials and reagents
Biological materials
1. ATCC Saccharomyces cerevisiae; W303-1a (ATCC, 50-238-3847)
2. Aetherobacter fasciculatus SBSr002 (Leibniz-Institute DSMZ-German Collection of Microorganisms and Cell Cultures, DSM 24601)
3. TransforMax EPI300 Electrocompetent E. coli (EpicentreTM, EC300110)
4. pCC1FOS (Epicentre Technologies, V008674)
5. pRS314 (ATCC, Plasmid #77143)
6. pRS315 (ATCC, Plasmid #77144)
7. pk18mobsacB (ATCC, Plasmid #87097)
8. E. coli DH5α competent cells (ThermoFisher Scientific, catalog number: 18265017)
Reagents
1. GenEluteTM Bacterial Genomic DNA kit (Sigma-Aldrich, catalog number: NA2120)
2. Agentcourt AMPure XP beads (Beckman Coulter, catalog number: A63880)
3. Rapid Barcoding kit (Oxford Nanopore Technologies, catalog number: SQK-RBK004)
4. Q5® high-fidelity DNA polymerase (New England BioLabs, catalog number: M0491S)
5. PhireTM Hot Start II DNA polymerase (Thermo Scientific, catalog number: F124S)
6. β-Mercaptoethanol (Millipore, catalog number: 444203)
7. Zymolyase-100T (Carl Roth, catalog number: 9329.1)
8. Vitamin B12 (Sigma-Aldrich, catalog number: V2876)
9. Yeast extract (Gibco, catalog number: 211931)
10. CaCl2 × 2H2O (Sigma-Aldrich, catalog number: 223506)
11. Agar (Difco, catalog number: 214530)
12. Sorbitol (Millipore, catalog number: 56755-M)
13. EDTA (Sigma-Aldrich, catalog number: EDS-100G)
14. Bacto peptone (Gibco, catalog number: DF0118-17-0)
15. Tris HCl (Roche, catalog number: 10812846001)
16. Na2HPO4 × 2H2O (Sigma-Aldrich, catalog number: 71643)
17. NaH2PO4 × 2H2O (Sigma-Aldrich, catalog number: 71505)
18. PEG 8000 (Fisher Scientific, catalog number: BP233-1)
19. Minimal SD base (Takara, catalog number: 630411)
20. Yeast synthetic drop-out medium supplement, without leucin, DO-Leu (Takara, catalog number: 630414)
21. NaOH (Supelco, catalog number: 1091401,000)
22. Sodium dodecyl sulfate (Sigma-Aldrich, catalog number: 436143)
23. YPDA (Takara, catalog number: 630464)
24. Yeast DNA Extraction kit (Thermo Scientific, catalog number: 78870)
25. 1 kb Plus DNA ladder (New England Biolabs, catalog number: N3200S)
26. Gel loading dye, purple (New England Biolabs, catalog number: B7025)
27. Gentamycin sulfate salt (BioReagent, catalog number: G1264)
28. Glycerol (Sigma-Aldrich, catalog number: 49767)
29. Chloramphenicol (Sigma-Aldrich, catalog number: C0378)
30. Buffer TAE (Sigma-Aldrich, catalog number: 574797)
31. KOH (Sigma-Aldrich, catalog number: P1767)
32. Oligonucleotides for amplification and colony PCR primers (Integrated DNA Technologies), storage: -20 °C
33. MIDORI Green Easy (Nippon genetics, catalog number: MG12)
34. NucleoSpin Gel and PCR Clean-up (Macherey-Nagel, catalog number: 740609.50)
35. GeneJET Plasmid Miniprep kit (Thermo Scientific, catalog number: K0502)
36. CHEF Genomic DNA Plug kits (Bio-Rad, catalog number: 170-3592)
37. HpaI (New England Biolabs, catalog number: R0105S)
38. BamHI-HF (New England Biolabs, catalog number: R3136S)
39. NaCl (Sigma-Aldrich, catalog number: S9625)
40. Tryptone (OXOID, catalog number: LP0042)
41. Baker’s yeast (Bruggeman, catalog number: 2001-0580)
Solutions
1. VY/2 media (see Recipes)
2. EDTA 0.5 M pH 7.5 (see Recipes)
3. Sorbitol 1 M (see Recipes)
4. SOS (see Recipes)
5. Tris-HCl 1 M pH 7.5 (see Recipes)
6. SPEM pH 7.5 (see Recipes)
7. STC (see Recipes)
8. PEG (see Recipes)
9. Zymolyase stock (see Recipes)
10. TOP agar-leu (or -Trp) (see Recipes)
11. Sorb plates -Leu (or -Trp) (see Recipes)
12. Liquid media YPDA (see Recipes)
13. SDS (see Recipes)
14. Reaction setup for Q5® High-Fidelity DNA Polymerase (see Recipes)
15. Reaction setup for PhireTM Hot Start II DNA Polymerase (see Recipes)
16. Gentamycin (see Recipes)
17. CaCl2 1 M (see Recipes)
18. Golden Gate assembly mixture (see Recipes)
19. Chloramphenicol (see Recipes)
20. KOH (see Recipes)
21. Glycerol (see Recipes)
22. LB medium (see Recipes)
Recipes
1. VY/2 media
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Baker’s yeast | 5 g/L | 5 g |
| CaCl2 × 2H2O | 1.36 g/L | 1.36 g |
| Vitamin B12 | 0.5 mg/L | 0.5 mg |
| Agar (Difco) | 15 g/L | 15 g |
| Distilled water | n/a | 1,000 mL |
Note: Autoclave. Adjust to pH 7.2 with KOH.
2. EDTA 0.5 M pH 7.5
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| CaCl2 | 0.18612 g/mL | 46.53 g |
| Distilled water | n/a | 250 mL |
Note: Filter sterilize the solution using a 0.22 μm sterile filter.
3. Sorbitol 1 M
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 182 g/L | 182 g |
| Distilled water | n/a | 1,000 mL |
Note: Filter sterilize the solution using a 0.22 μm sterile filter. Storage: 6 months at room temperature (RT).
4. SOS
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 0.182 g/mL | 9.1 g |
| Yeast extract | 0.0025 g/mL | 0.125 g |
| Bacto peptone | 0.005 g/mL | 0.25 g |
| CaCl2 | 6.5 mM | 300 μL |
| Distilled water | n/a | 50 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter. Store for 6 months at RT. Aliquot in 5 mL volumes.
5. Tris-HCl 1 M pH 7.5
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris-HCl 1 M pH 7.5 | 121.4 g/L | 121.4 g |
| Distilled water | n/a | 1,000 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter.
6. SPEM pH 7.5
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Na2HPO4 × 2H2O | 0.00138 g/mL | 0.69 g |
| NaH2PO4 × 2H2O | 0.00036 g/mL | 0.18 g |
| EDTA pH 7.5 | 0.5 M | 10 mL |
| Sorbitol 1 M | 0.182 g/mL | 91 g |
| Distilled water | n/a | 500 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter. Store for 6 months at RT. Aliquot in 10 mL volumes.
7. STC
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 0.182 g/mL | 18.2 g |
| Tris-HCl 1 M pH 7.5 | 0.01 M | 1 mL |
| CaCl2 1 M | 0.01 M | 1 mL |
| Distilled water | n/a | 100 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter. Store for 6 months at RT. Aliquot in 1 mL volumes.
8. PEG
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| PEG 8000 | 20% | 4 g |
| Tris-HCl 1 M pH 7.5 | 10 mM | 200 μL |
| CaCl2 1 M | 10 mM | 200 μL |
| Distilled water | n/a | 20 mL |
Note: Heat mildly if necessary. Filter sterilize the solution using a 0.2 μm sterile filter. Prepare fresh.
9. Zymolyase stock
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Zymolyase-100T | 10 mg/mL | 200 mg |
| Tris-HCl 1 M pH 7.5 | 0.1 M | 1 mL |
| Glycerol 50% | 5% | 10 mL |
| Distilled water | n/a | 9 mL |
Note: Aliquot and store for 6 months at -20 °C.
10. TOP agar-leu (or -Trp)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol 1 M | 0.182 g/mL | 91 g |
| SD base | 0.0267 g/mL | 13.35 g |
| DO-Leu (or -Trp) | 0.0007 g/mL | 0.35 g |
| Agar | 0.03 g/mL | 15 g |
| Distilled water | n/a | 500 mL |
Note: Autoclave and store at 65 °C. Adjust to pH 5.8 with NaOH 1 M.
11. Sorb plates -Leu (or -Trp)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 1 M | 145.6 g |
| SD base | 0.0267 g/mL | 21.36 g |
| DO-Leu (or -Trp) | 0.0006875 g/mL | 0.55 g |
| Agar | 0.02 g/mL | 16 g |
| Distilled water | n/a | 800 mL |
Note: Autoclave and store the Sorb agar bottle at 65 °C. Adjust to pH 5.8 with NaOH 1 M.
12. Liquid media YPDA
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| YPDA | 1× | 25 g |
| Distilled water | n/a | 500 mL |
Note: Autoclave.
13. SDS
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| SDS | 0.02 g/mL | 2 g |
| Distilled water | n/a | 100 mL |
14. Reaction setup for Q5® High-Fidelity DNA Polymerase
| Reagent | Final concentration | Quantity or volume for 25 μL reaction |
|---|---|---|
| 5× Q5® reaction buffer | 1× | 5 μL |
| 10 mM dNTPs | 200 μM | 0.5 μL |
| 10 μM forward primer | 0.5 μM | 1.25 μL |
| 10 μM reverse primer | 0.5 μM | 1.25 μL |
| Template DNA | <1,000 ng | Variable (usually 0.25 µL) |
| Q5® High-Fidelity DNA Polymerase | 0.02 U/µL | 0.25 μL |
| 5× Q5® High GC Enhancer | 1× | 5 μL |
| Nuclease-free water | - | to 25 μL |
15. Reaction setup for PhireTM Hot Start II DNA Polymerase
| Reagent | Final concentration | Quantity or volume for 20 μL reaction |
|---|---|---|
| 5× Phire Green reaction buffer | 1× | 4 μL |
| 10 mM dNTPs | 200 μM | 0.4 μL |
| 10 μM forward primer | 0.5 μM | 1 μL |
| 10 μM reverse primer | 0.5 μM | 1 μL |
| Template DNA | - | Variable |
| PhireTM Hot Start II DNA Polymerase | - | 0.4 μL |
| DMSO | 1× | 0.6 μL |
| Nuclease-free water | - | Add to 20 μL |
16. Gentamycin
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Gentamycin sulfate salt | 15 μg/mL | 150 μg |
| Distilled water | n/a | 10 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter. Store at -20 °C. Aliquot in 1 mL volumes.
17. CaCl2 1 M
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| CaCl2 | 0.111 g/mL | 1.11 g |
| Distilled water | n/a | 10 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter.
18. Golden Gate assembly mixture
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| BsaI-HF® v2 | 10 μL | |
| T4 DNA ligase | 10 μL | |
| T4 buffer | 15 μL | |
| BSA | 1.5 μL | |
| MilliQ water | 13.5 μL |
19. Chloramphenicol
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Chloramphenicol | 35 μg/mL | 500 mg |
| 95% ethanol | n/a | 14.28 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter. Store at -20 °C. Aliquot in 1 mL volumes.
20. KOH
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Potassium hydroxide | 0.1 M | 5.6 g |
| Distilled water | n/a | 1,000 mL |
Note: Filter sterilize the solution using a 0.2 μm sterile filter. Store at -20 °C. Aliquot in 1 mL volumes.
21. Glycerol
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Glycerol | 50% | 25 mL |
| Distilled water | n/a | 25 mL |
Note: Autoclave.
22. LB medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tryptone | 10 g/L | 10 g |
| NaCl | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| Distilled water | n/a | 1,000 mL |
Note: Autoclave.
Laboratory supplies
1. Vented cap Greiner tubes (Greiner bio-one, catalog number: 115262)
2. UV cuvettes (Brand, catalog number: Z628026)
3. 10 μL pipette tips (Brand 732724, catalog number: Z740051)
4. 20 μL pipette tips (Brand 732724, catalog number: Z740051)
5. 200 μL pipette tips (Brand 732724, catalog number: Z740051)
6. 1,000 μL pipette tips (Brand 732724, catalog number: Z740051)
7. Erlenmeyer flasks 500 mL (Duran, catalog number: 212174405)
8. 1.5 mL microcentrifuge tube (Brand, catalog number: 525-0944)
9. Falcon tubes 50 mL (Corning, catalog number: 352098)
10. 0.2 mL thin-walled PCR tubes (VWR, catalog number: 732-1521)
11. Gene Pulser/MicroPulser electroporation cuvettes, 0.2 cm gap (Bio-Rad, catalog number: 1652082)
12. 0.22 μm syringe filter (Millipore, catalog number: SLGPR33RS)
13. Syringe PP/PE without needle (Sigma-Aldrich, catalog number: Z683604)
Equipment
1. Microfuge (Benchmark Scientific C1008-R-E-UK, catalog number: Z764205)
2. Microcentrifuge (Eppendorf, catalog number: 5406000119)
3. Pipette 2–20 μL (Thermofisher, catalog number: 4642060)
4. Pipette 20–200 μL (Thermofisher, catalog number: 4642080)
5. Pipette 100–1,000 μL (Thermofisher, catalog number: 4642090)
6. Freezer (-20 °C)
7. Refrigerator (2–8 °C)
8. Benchtop centrifuge (Thermo Scientific, catalog number: 28523)
9. Incubator (FED, catalog number: 260)
10. Incubator shaker (New Brunswick Innova®, catalog number: M1335-0002)
11. Thermomixer (Eppendorf, catalog number: EP5387000030)
12. OD meter 600 nm (Implen DiluphotometerTM, catalog number: C40)
13. Nanodrop (Thermofisher, catalog number: ND-2000)
14. MinION Flow Cell R9.4.1 (Oxford Nanopore Technologies, catalog number: FLO-MIN106D)
15. MinION (Oxford Nanopore Technologies, M1DCapEx)
16. Electronic Pipette Controller (Eppendorf® Easypet®, catalog number: EP4430000018)
17. Fume hood
18. Laminar flow hood
19. Mupid-One Electrophoresis Unit (MUPIDONE, catalog number: MU2)
20. Gel casting tray (MUPIDONE, catalog number: ON-MS)
21. Well combs (MUPIDONE, catalog number: AC-C1)
22. Mastercycler Nexus X2 Thermal Cyclers Nexus GX2e satellite unit (Eppendorf, catalog number: EP6338000012)
23. Gel Doc XR+ (Bio-Rad, catalog number: 170-8195)
24. Gene Pulser Xcell Electroporation Systems (Bio-Rad, catalog number: 1652660)
25. pH meter (Mettler Toledo, catalog number: MT30266626)
26. Hula mixer (rotator mixer) (Thermo Scientific, catalog number: 15920D)
Software and datasets
1. Guppy Oxford Nanopore Technologies 4.4.1(https://github.com/nanoporetech/pyguppyclient)
2. Flye genome assembler v2.9.5 (https://github.com/mikolmogorov/Flye)
3. Medaka polisher v1.12.0 (https://github.com/nanoporetech/medaka).
4. antiSMASH v7.1 (https://github.com/antismash/antismash/releases)
5. checkM v1.2.3 (https://github.com/Ecogenomics/CheckM)
6. BUSCO 5.6.0 (https://gitlab.com/ezlab/busco)
7. Bakta v1.9.3 (https://github.com/oschwengers/bakta).
8. COG classifier tool v1.0.5 (https://github.com/moshi4/COGclassifier).
All data have been deposited in the European Nucleotide Archive under the accession PRJEB72099 and Campos-Magaña et al. [18].
Procedure
文章信息
稿件历史记录
提交日期: Sep 24, 2025
接收日期: Nov 7, 2025
在线发布日期: Nov 27, 2025
出版日期: Dec 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Campos-Magaña, M. A., Martins dos Santos, V. A. P. and Garcia-Morales, L. (2025). A Rapid and Cost-Effective Pipeline to Identify and Capture BGCs From Bacterial Draft Genomes. Bio-protocol 15(24): e5549. DOI: 10.21769/BioProtoc.5549.
分类
微生物学 > 微生物遗传学 > 全基因组测序
分子生物学 > DNA > DNA 克隆
生物信息学与计算生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link









