- EN - English
- CN - 中文
Synchronizing Germination Rates Across Plant Species for Fabricated Ecosystems EcoFAB 2.0
在 EcoFAB 2.0 人工生态系统中实现多种植物萌发速率的同步
发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5537 浏览次数: 1448
评审: Samik BhattacharyaShweta PanchalAnonymous reviewer(s)

相关实验方案
利用体外渗透梯度实验系统评估拟南芥主根对渗透胁迫的响应生长
Selene Píriz-Pezzutto [...] Mariana Sotelo-Silveira
2025年07月20日 1902 阅读
Abstract
Roots are essential organs for plants, facilitating water and nutrient uptake from the soil to support growth. Traditional methods for studying root systems, such as rhizoboxes and rhizotrons, have provided valuable insights. However, advanced methods such as fabricated ecosystems (EcoFAB) combined with new generation microscopes now enable a more detailed investigation of the rhizosphere, the microenvironment surrounding roots, allowing a deeper understanding of root tissue, exudates, and plant–soil interactions. This microenvironment can be used to investigate the adaptation of plants to environmental stress (salinity, drought, higher temperatures). Our procedure focuses on establishing standardized protocols for plant growth tailored to the EcoFAB system, which offers a controlled environment to study root dynamics. This work also contributes new insights into the early stages of plant germination, an area currently underexplored in the literature. While numerous studies focus on plant growth or genetic aspects, such as gene induction, the germination phase remains underexplored. We have developed optimized germination protocols for multiple plant species, ensuring uniform seedling size and sufficient development for seamless integration into the EcoFAB system.
Key features
• Optimized the germination protocol for plant rhizosphere studies using the EcoFAB 2.0 system.
• Adapted the well-established protocol for Brachypodium distachyon to other species, including Arabidopsis thaliana, Fragaria vesca, and Lactuca sativa.
• Preliminary measurements of root and leaf traits were taken to determine the best method.
Keywords: Fabricated ecosystem (人工生态系统)Graphical overview
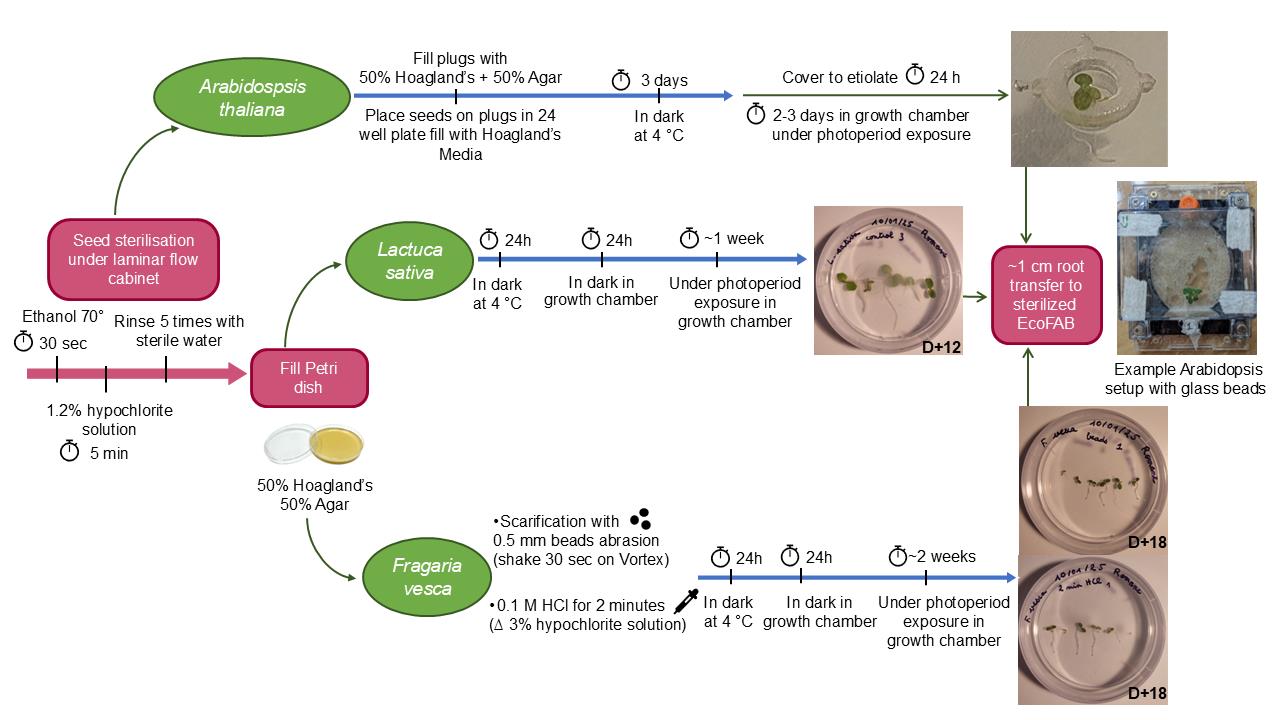
Seed preparation procedure and initial growth prior to transfer to fabricated ecosystems EcoFAB 2.0 for Arabidopsis thaliana, Lactuca sativa, and Fragaria vesca.
Background
The understanding of root development and its surrounding environment, the rhizosphere, under varying conditions is crucial for optimizing plant growth and yield for agriculture. The study of plant physiological processes in response to their environment is possible with fabricated ecosystems (EcoFAB 2.0 system). This system allows high-resolution imaging of roots in a multi-factorial, controlled environment [1,2] and the characterization of shoot growth. EcoFABs enable the study of plant–microbiome interactions by introducing microbial, fungal, or bacterial cultures into the chamber, which can be easily sterilized. During experiments, the nutrient media can be replaced or altered at any time. Imaging or scanning can be conducted to collect data on root size, root hairs, hyphae, leaves, and interparticle space. Root architecture and leaf growth can be analyzed simultaneously. By imposing different conditions on the plants, we can observe which ones encourage or hinder their development.
The development of the EcoFAB system, a repeatable and versatile tool for studying plants in controlled environments, enables a more effective transfer of knowledge across different scientific communities. A standardized sterilization and germination protocol for various plant species is necessary to ensure uniform plant growth stages suitable for transfer into the sterilized EcoFAB system. Seed priming allows the control of seed hydration and induces physiological, biochemical, and molecular alterations that trigger germination [3]. With this type of treatment, it is possible to increase vigor and germination rate or reduce the time needed to obtain better seedlings. Radicle protrusion through the seed coat is the main event to observe homogeneous germination. To enhance germination, many invasive methods [4,5] have been described in the literature, mainly based on water uptake by seeds. This is why we developed a user-friendly seed treatment and germination protocol for different plant species to achieve a homogeneous germination rate and growth suitable for experiments in the EcoFAB system.
Several other tools are available to study plants and their interactions with soil, microorganisms, and ecosystems. Systems like rhizoboxes and rhizotrons [6] were initially created to study root growth in mature plants. These systems enable plants to grow in their natural soil environment. Using a non-destructive method, root or shoot growth can be assessed through successive measurements on the same plants. This approach reduces phenotypic variability linked to differences between individual plants and enhances accuracy compared to methods requiring plant destruction during sampling [7]. Recently, the CD-Rhizotron system [8] was developed to study the spatial differentiation of roots linked to different microbial communities or exudates. Fine root studies are also possible with Minirhizotrons [9] or EnRoot [10] devices, but as with previous methods, the ecosystem is not controlled, and plants remain in the field. More recent technologies, such as TRIS [11] and root arrays [12], have been developed to achieve a precise understanding of root behavior in microfluidic devices, imaging them in real-time and enabling genotyping analysis. A key limitation of these devices is their restriction to experiments on small, young plants. Additionally, these systems do not replicate field-like environments.
In contrast, EcoFABs are fully controllable hydroponics systems, offering many possibilities for experiments. A deep understanding of plant growth mechanisms and interactions has numerous direct applications in agriculture, such as improving yield and triggering some diseases caused by fungi or bacteria. Conservation efforts for endangered species are another important application, as the International Union for Conservation of Nature (IUCN) red list encourages the scientific community to engage in conservation efforts for these species. Early developmental stages are crucial for the conservation of endangered species, and EcoFABs could help determine key features of early development, particularly by optimizing life cycles and seed germination. Beyond these applications, studying plant–microbiome–soil interactions in systems like the EcoFAB holds significant potential for space exploration. As part of the Plants 4 Space Program, this research explores the use of EcoFABs in optimizing fruit and vegetable production for long-term space missions. A deep understanding of root cells and rhizosphere interactions is essential for optimizing crops in space [13], especially fresh food production to counteract the degradation of packaged food and the potential digestive issues caused by a lack of fruits and vegetables in astronauts’ diets [14]. This work introduces the potential use of strawberries [15] and lettuce in EcoFAB 2.0, aligning with space programs that have mainly focused on these crops. It also opens the door to studying additional plants in the EcoFAB system.
Inspired by the Brachypodium distachyon protocol [16,17], we present a precise germination and setup protocol in EcoFABs for Arabidopsis thaliana with the aim of extending the study of this model plant. Lettuce is widely studied in hydroponic systems for vegetable production [18,19]. Here, we developed a new approach to studying lettuce (Lactuca sativa; Outredgeous) in its early stages of development. We also propose a quite similar protocol applicable for Fragaria vesca (Hawaii 4). Our protocol can be easily adapted to many other species, including endangered ones. The main goal is to extend EcoFAB 2.0's applicability to additional plant species and provide standardized procedures that improve both the quantitative (germination rate) and qualitative (uniformity) aspects of plant germination.
Materials and reagents
Biological materials
1. Arabidopsis seeds (Columbia, wild-type seeds from the Arabidopsis Biological Resource Center at The Ohio State University; seeds were bulked up from that stock in our lab)
2. Lactuca sativa “Outredgeous” seeds (The Seed collection, Z-10139_P)
3. Fragaria vesca “Hawaii 4” seeds (original seeds from https://npgsweb.ars-grin.gov/gringlobal/accessiondetail?id=1856850; seeds were bulked up from that stock in our lab)
Reagents
1. Plant cell culture agar powder (Sigma-Aldrich, catalog number: A1296-5KG)
2. 100% ethanol (Bio21, catalog number: EA043-2.5L-J)
3. Sodium hypochlorite solution 12% (Thermo Fisher Scientific, catalog number: AJA485-5L)
4. TritonTM X-100 (Sigma-Aldrich, catalog number: X100-100ML)
5. Hydrochloric acid (HCl), 37% (Sigma-Aldrich, catalog number: 258148-500ML, CAS 7647-01-0)
6. Potassium nitrate (KNO3) (Sigma-Aldrich, CAS number: 7757-79-1)
7. Ammonium dihydrogen phosphate [(NH4)2HPO4] (Sigma-Aldrich, CAS number: 7783-28-0)
8. Calcium nitrate tetrahydrate [Ca(NO3)2·4H2O] (Sigma-Aldrich, CAS number: 13477-34-4)
9. Magnesium sulphate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, CAS number: 10034-99-8)
10. Boric acid (H3BO3) (Sigma-Aldrich, CAS number: 10043-35-3)
11. Manganese chloride tetrahydrate (MnCl2·4H2O) (Sigma-Aldrich, CAS number: 13446-34-9)
12. Zinc sulphate heptahydrate (ZnSO4·7H2O) (Sigma-Aldrich, CAS number: 744620-0)
13. Copper sulphate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, CAS number: 7758-99-8)
14. Sodium molybdate dihydrate (Na2MoO4·2H2O) (Sigma-Aldrich, CAS number: 10102-40-6)
15. Iron (II) sulphate heptahydrate (FeSO4·7H2O) (Sigma-Aldrich, CAS number: 7782-63-0)
16. EDTA disodium salt (C10H14N2Na2O8) (Sigma-Aldrich, CAS number: 6381-92)
Solutions
1. Sodium hypochlorite solution 1.2% (see Recipes)
2. Sodium hypochlorite solution 3% (see Recipes)
3. Hoagland’s nutrient solution [20]
a. Micronutrients solution (see Recipes)
b. Macronutrients solution (see Recipes)
c. Iron solution (see Recipes)
d. Final Hoagland’s nutrient solution (see Recipes)
Recipes
1. Sodium hypochlorite solution 1.2%
| Reagent | Final concentration | Volume |
|---|---|---|
| Sterile MilliQ water | 43 mL | |
| Triton X-100 | 0.106 g/mL | 2 mL |
| Sodium hypochlorite solution (12%) | 12 g/L | 5 mL |
| Total | 50 mL |
Prepare under sterile conditions in a laminar flow cabinet to minimize contamination. We recommend storing it at 4 °C and making it fresh every fortnight.
2. Sodium hypochlorite solution 3%
| Reagent | Final concentration | Volume |
|---|---|---|
| Sterile MilliQ water | 35.5 mL | |
| Triton X-100 | 0.106 g/mL | 2 mL |
| Sodium hypochlorite solution (12%) | 37.2 g//L | 15.5 mL |
| Total | 50 mL |
Prepare under sterile conditions in a laminar flow cabinet to minimize contamination. We recommend storing it at 4 °C and making it fresh every fortnight.
3. Hoagland’s nutrient solution
Create a collection of concentrated stock solutions for each ingredient that can be filter-sterilized. Dissolve the following salts in individual 50 mL aliquots of MilliQ water. Under a laminar flow, filter-sterilize each stock solution into autoclaved bottles. Store in the dark and at room temperature.
a. Macronutrient solutions (four separate solutions)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| KNO3 | 303.4 g/L | 15.17 g |
| NH4H2PO4 | 115 g/L | 5.75 g |
| Ca(NO3)2·4H2O | 944.6 g/L | 47.23 g |
| MgSO4·7H2O | 493 g/L | 24.65 g |
b. Micronutrient solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| H3BO3 | 2.86 g/L | 143 mg |
| MnCl2·4H2O | 3.76 g/L | 188.1 mg |
| CuSO4·5H2O | 0.13 g/L | 6.3 mg |
| ZnSO4·7H2O | 0.39 g/L | 19.6 mg |
| Na2MoO4·2H2O | 0.03 g/L | 1.3 mg |
c. Iron solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Fe(III)SO4·7H2O | 5.56 g/L | 278 mg |
| C10H14N2Na2O8 | 7.44 g/L | 372 mg |
Make sure to minimize light exposure of the iron solution by covering the autoclaved bottles with aluminum foil.
d. Final Hoagland’s nutrient solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sterile MilliQ water | 1 L | |
| KNO3 | 0.607 g/L | 0.607 g |
| NH4H2PO4 | 0.115 g/L | 0.115 g |
| Ca(NO3)2·4H2O | 0.995 g/L | 0.995 g |
| MgSO4·7H2O | 0.493 g/L | 0.493 g |
| H3BO3 | 2.8 mg/L | 2.8 mg |
| MnCl2·4H2O | 3.762 mg/L | 3.762 mg |
| CuSO4·5H2O | 0.392 mg/L | 0.392 mg |
| ZnSO4·7H2O | 0.126 mg/L | 0.126 mg |
| Na2MoO4·2H2O | 0.026 mg/L | 0.026 mg |
| Fe(III)SO4·7H2O | 5.56 mg/L | 5.56 mg |
| C10H14N2Na2O8 | 7.44 mg/L | 7.44 mg |
| Total | 1 L |
Hoagland’s nutrient solution is prepared by adding 2 mL of KNO3 and 1 mL of all other stock solutions [NH4H2PO4, Ca(NO3)2·4H2O, MgSO4·7H2O, micronutrient solution, and iron solution] to 991 mL of MilliQ water in an autoclaved bottle under sterile conditions in a laminar flow cabinet. The pH is adjusted to 6 mainly so that plants and microorganisms can also grow in this media. This yields a sterile Hoagland’s nutrient solution with the composition described above. Store it at room temperature.
Laboratory supplies
1. 24-well tissue culture treated plates, with lid, flat bottom, sterile (Adelab Scientific, catalog number: CNG3526; referred to as 24-well plates)
2. Petri dish 90 × 25 mm (Thermo Fisher Scientific, catalog number: LBS60014X)
3. Petri dish 90 × 15 mm (Thermo Fisher Scientific, catalog number: LBS60001)
4. Plugs, made by Lawrence Berkeley National Laboratory in collaboration with us; size-V3 plugs worked best (more information can be found at https://eco-fab.org/)
5. Pipette tips 100–1,000 μL (Mettler-Toledo, catalog number: 17014969)
6. Pipette tips 100–200 μL (Mettler-Toledo, catalog number: 17014964)
7. Falcon® 50 mL, conical bottom, sterile (Bio-Strategy Pty Limited, catalog number: 352070)
8. Eppendorf tube 2 mL (Eppendorf, catalog number: 0030120094)
9. Borosilicate beads 0.5 mm diameter (Merck, catalog number: Z250465)
10. Forcep jeweller (McFarlane Medical, catalog number: 34288-LQ)
11. Micropore tape (3M, catalog number: 1530-0)
12. Aluminum foil
13. EcoFAB 2.0 (more information can be found at https://eco-fab.org/)
14. Screwdriver (Panasonic, EY 7412 SB, Cordless Screw Driver)
Equipment
1. Laminar flow cabinet (Clyde-Apac, model: HWS120)
2. Vortex mixer (Ratek, model: VM1)
3. Growth chamber (Bioline Global, model: AR-22L)
4. Freezer
5. Autoclave
6. EVOS Microscope (Thermo Fisher Scientific, model: EVOS M7000 Imaging System)
7. WinRHIZO Scanning system (Regent Instruments)
Software and datasets
1. CellesteTM 5 Image Analysis Software (Thermo Fisher Scientific, AMEP4877) or Fiji image analysis, free software package available on GitHub
Procedure
文章信息
稿件历史记录
提交日期: Aug 20, 2025
接收日期: Oct 30, 2025
在线发布日期: Nov 18, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Charbeaux, R. S. F., Waymouth, V. J., Calabria, J., Miller, T., Andeer, P. and Watt, M. (2025). Synchronizing Germination Rates Across Plant Species for Fabricated Ecosystems EcoFAB 2.0. Bio-protocol 15(23): e5537. DOI: 10.21769/BioProtoc.5537.
分类
植物科学 > 植物生理学 > 植物生长
植物科学 > 植物生理学 > 非生物胁迫
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link