- EN - English
- CN - 中文
Imaging the Entire Sexual Life Cycle of the Budding Yeast Saccharomyces cerevisiae Using a Microfluidic Platform
利用微流控平台成像观察酿酒酵母整个有性生殖周期
(*Contributed equally to this work, §Technical contact: tkenned2@ncsu.edu) 发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5536 浏览次数: 1477
Abstract
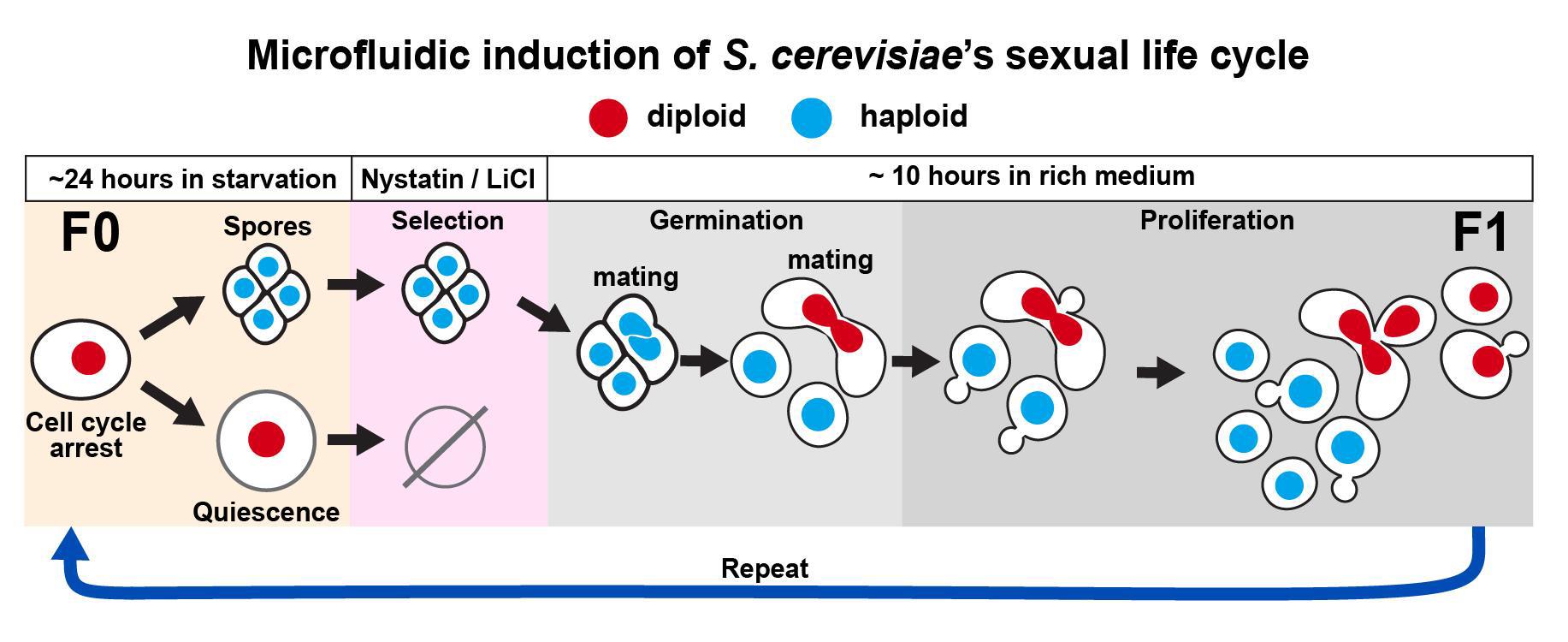
Microbial life cycles are often reconstructed theoretically from fragmentary pieces of evidence. Protocols for the direct and continuous observation of entire microbial life cycles, including sexual reproduction, are scarce, which limits the study of cellular transitions between different life cycle stages and prevents the visualization of cryptic stages. Although sequence-based techniques, such as -omics approaches, can reconstruct cellular transitions at the genetic and biochemical level, these methods are destructive and do not recover information from the same living cell over time. This protocol provides a solution to directly and continuously observe microbial life cycles, including sexual reproduction, by using microfluidics manipulations that expose single cells to nutritional stimuli and selective pressures. As proof of principle, we triggered a life cycle sequence transition in the model yeast Saccharomyces cerevisiae, starting with an arrest of proliferation in an ancestor cell followed by induction of meiosis through starvation, selection of sexually reproducing cells through exposure to a drug cocktail, germination of haploid spores, and mating of haploid individuals, creating a new descendant generation. This protocol offers the possibility to directly compare molecular and cellular behavior across life cycle stages and across sexually reproducing generations.
Key features
• Outlines optimal microfluidic conditions for all life cycle stages in the model microorganism S. cerevisiae.
• Describes a selection method for sexually reproducing sporulated yeast cells and against quiescent cells.
• Provides descriptions for optimal loading of yeast cells in microfluidic devices for long-term imaging.
• Enables continuous detection across life cycle transitions, which could be applied to other biomedically and agriculturally relevant fungal microorganisms.
Keywords: Microbial life cycle (微生物生命周期)Graphical overview
Background
The life cycle of Saccharomyces cerevisiae has been used as a model system for studying all major transitions in eukaryotic organisms, ranging from cell division to sexual reproduction [1–3]. Historically, each life cycle stage has mostly been studied separately, and direct comparisons between life cycle stages are usually not possible because of different experimental requirements. For instance, protocols for studying meiosis require the use of synchronization methods, which affect cell cycle pathways, preventing a direct comparison of different cell states with minimal perturbations. As a result, descriptions of the yeast life cycle largely rely on fragmentary genetic and microscopy evidence instead of direct and continuous observations, a problem that is also prevalent in other biomedically and ecologically important microorganisms [4,5].
Directly observing complete life cycles can offer a solution to problems such as confusing life cycle stages from the same organism as different species [6,7] and the incorrect identification of sexual stages [8], and could help reveal cryptic microbial life cycle transitions [4,7].
A solution to directly observe life cycle transitions is to use microfluidic devices coupled to time-lapse microscopy, which has been used to directly visualize entire asexual bacterial [9,10] and eukaryotic [11] life cycles in living single cells. However, most experiments deal with short-term processes, especially since the long-term imaging of cells requires experimental conditions for achieving complex cellular transitions that can take days.
To enable long-term imaging of life cycle transitions, we designed this protocol to induce the entire sexual life cycle of S. cerevisiae, from proliferation into meiosis, germination, mating, and the proliferation of descendant cells across generations of sexually reproducing diploids. We used a commercially available microfluidic device and a combination of media and micro-culturing conditions that promote transitions across each stage of the yeast life cycle, with minimal perturbation to single cells, and for up to three sexually reproducing generations [12].
This protocol has the potential to enable a complete characterization of the life cycle of yeast cells and to quantitatively study other microbial life cycles, which is crucial to understanding ecological and evolutionary processes in microbial parasites and pathogens [13,14].
Materials and reagents
Biological materials
1. Saccharomyces cerevisiae Meyen ex E.C. Hansen 204722TM ATCC SK1 MATa/MATalpha HO can1(r) gal2 cup(s). Wild type prototroph (HIS3/HIS3, TRP1/TRP1, LEU2/LEU2, URA3/URA3) [15]
Reagents
1. Adenine hydrochloride hemihydrate (Tokyo Chemical Industry Co., LTD., CAS number: 2922-28-3)
2. Uracil (Tokyo Chemical Industry Co., LTD., CAS number: 66-22-8)
3. L-Histidine (Spectrum Chemical MFG Corp, catalog number: H1021, CAS number: 71-00-1)
4. L-Leucine (Research Products International, catalog number: L22000-100.0, CAS number: 61-90-5)
5. L-Tryptophan (Tokyo Chemical Industry Co., LTD., CAS number: 73-22-3)
6. Arginine hydrochloride (Spectrum Chemical MFG Corp, catalog number: A1337, CAS number: 1119-34-2)
7. L-Aspartic acid (Sigma-Aldrich, catalog number: A9256-100G, CAS number: 56-84-8)
8. L-Glutamic acid (Tokyo Chemical Industry Co., LTD., CAS number: 56-86-0)
9. L-Phenylalanine (Tokyo Chemical Industry Co., LTD., CAS number: 63-91-2)
10. L-Lysine hydrochloride (Research Products International, catalog number: L37040-500.0, CAS number: 657-27-2)
11. L-Methionine (Sigma-Aldrich, catalog number: M5308-25G, CAS number: 63-68-3)
12. L-Serine (Thermo Scientific Chemicals, catalog number: 132661000)
13. L-Threonine (Research Products International, CAS number: 72-19-5)
14. L-Valine (Tokyo Chemical Industry Co., LTD., CAS number:72-18-4)
15. L-(-)-Tyrosine (Tokyo Chemical Industry Co., LTD., CAS number: 60-18-14)
16. L-Isoleucine (Tokyo Chemical Industry Co., LTD., CAS number:73-32-5)
17. Lithium chloride (LiCl) (Sigma-Aldrich, catalog number: L9650-500G, CAS number: 7447-41-8)
18. D(+)-Glucose, ACS reagent, anhydrous (Thermo Scientific Chemicals, catalog number: 410950050, CAS number: 50-99-7)
19. Succinic acid (Research Products International, catalog number: S42000, CAS number: 110-15-6)
20. Sodium hydroxide (Spectrum Chemical MFG Corp, CAS number: 1310-73-2)
21. Ammonium sulfate (Research Products International, catalog number: A20510, CAS number: 7783-20-2)
22. Yeast nitrogen base without amino acids and ammonium sulfate (Sigma-Aldrich, catalog number: Y1251)
23. BactoTM yeast extract (Gibco, catalog number: DF0127-17-9)
24. BactoTM peptone (Gibco, catalog number: 211677)
25. Agar, powdered bacteriological grade (Apex BioResearch Products, catalog number: 20-273)
26. Potassium acetate (Research Products International, catalog number: P42025-500.0, CAS number: 127-08-2)
27. Nystatin, powder (Sigma, CAS number: 1400-61-9)
28. Sodium carbonate (Na2CO3) (Spectrum Chemical MFG Corp., catalog number: S1226, CAS number: 497-19-8)
29. Sorbitol (Sigma-Aldrich, catalog number: S1876-1KG)
30. Glycerol (Sigma-Aldrich, catalog number: G9012-2L, CAS number: 56-81-5)
31. Dimethyl sulfoxide (DMSO) (Fisher Scientific, catalog number: BP231-100)
Solutions
1. 4 M LiCl solution (see Recipes)
2. 20% glucose solution (see Recipes)
3. 2% sorbitol solution (see Recipes)
4. 0.25 M Na2CO3 solution (see Recipes)
5. Synthetic complete dropout powder (see Recipes)
6. Synthetic complete dextrose liquid medium (SCD) (see Recipes)
7. YPD liquid media (see Recipes)
8. YPD plates (see Recipes)
9. Life cycle sporulation liquid media (LC-SPO) (see Recipes)
10. Stock Nystatin solution in DMSO (see Recipes)
Recipes
1. 4 M LiCl solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Lithium chloride | 4 M | 17.0 g |
| diH2O | – | 100 mL (fill up to) |
| Total | n/a | 100 mL |
Prepare 100 mL of 4 M LiCl stock solution by adding 17.0 g of LiCl powder to a sterile 100 mL bottle containing 50 mL of sterile deionized water (diH2O). Allow the LiCl to fully dissolve and bring the final volume to 100 mL with sterile diH2O. Store at room temperature.
2. 20% glucose solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Glucose | 20% | 200 g |
| diH2O | – | 1,000 mL (fill up to) |
| Total | n/a | 1,000 mL |
Prepare 1 L of stock 20% glucose solution by adding 200 g of glucose powder to a 1 L flask or beaker containing 500 mL diH2O, stirring with a stir bar on a stir plate. Allow the glucose to fully dissolve, remove the stir bar, and bring the final volume to 1 L with diH2O. Filter-sterilize using a 0.2 μm Nalgene® bottle-top sterile connected to a vacuum line. See sterilization protocols in Table 1.
Table 1. Sterilization protocols
| Solution | Sterilization |
| 20% glucose; 2% sorbitol | Filter-sterilize using a 0.2 μm pore size Nalgene® bottle-top sterile filter connected to a vacuum line. |
| YPD liquid (without glucose) | Autoclave using the 30-min liquid cycle in PRIMUS and Amsco autoclave systems: 121 °C, 15 psi, 30 min. |
| YPD agar solid (without glucose) | |
| LC-SPO (without Na2CO3) |
3. 2% Sorbitol solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 2% | 20 g |
| diH2O | – | 1,000 mL (fill up to) |
| Total | n/a | 1,000 mL |
Prepare 1 L of 2% sorbitol solution by adding 20 g of sorbitol powder to a 1 L flask or beaker containing 500 mL of diH2O, stirring with a stir bar on a stir plate. Allow the sorbitol to fully dissolve, remove the stir bar, and bring the final volume to 1 L with diH2O. Filter-sterilize using a 0.2 μm Nalgene® bottle-top sterile connected to a vacuum line (see Table 1).
4. 0.25 M Na2CO3 solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Na2CO3 | 0.25 M | 2.65 g |
| diH2O | – | 100 mL (fill up to) |
| Total | n/a | 100 mL |
Prepare 100 mL of 0.25 M Na2CO3 solution by adding 2.65 g of Na2CO3 powder to a sterile 100 mL bottle containing 50 mL of sterile diH2O. Allow the Na2CO3 to fully dissolve and bring the final volume to 100 mL with sterile diH2O. Store at room temperature.
5. Synthetic complete dropout powder
| Reagent | Final concentration | Quantity |
|---|---|---|
| Adenine hydrochloride hemihydrate | 2.91% | 1.6 g |
| Uracil | 1.40% | 0.8 g |
| L-Histidine | 1.40% | 0.8 g |
| L-Leucine | 8.70% | 4.8 g |
| L-Tryptophan | 5.80% | 3.2 g |
| Arginine hydrochloride | 1.40% | 0.8 g |
| L-Aspartic acid | 7.20% | 4.0 g |
| L-Glutamic acid | 7.20% | 4.0 g |
| L-Phenylalanine | 3.60% | 2.0 g |
| L-Lysine hydrochloride | 2.20% | 1.2 g |
| L-Methionine | 1.40% | 0.8 g |
| L-Serine | 27.10% | 15.0 g |
| L-Threonine | 14.40% | 8.0 g |
| L-Valine | 10.80% | 6.0 g |
| L-(-)-Tyrosine | 2.20% | 1.2 g |
| L-Isoleucine | 2.20% | 1.2 g |
| Total | n/a | 55.4 g |
Weigh, combine, and grind the powders of the amino acids described in the table with a clean mortar and pestle until the powder mixture is homogeneously white. Store at room temperature and away from sunlight in an amber bottle.
6. Synthetic complete dextrose liquid media (SCD)
| Reagent | Final concentration | Quantity or volume (for 1 L) |
|---|---|---|
| Succinic acid | 10 g/L | 10 g |
| Sodium hydroxide | 6 g/L | 6 g |
| Ammonium sulfate | 5 g/L | 5 g |
| Yeast nitrogen base without amino acids and ammonium sulfate | 1.7 g/L | 1.7 g |
| Complete dropout powder | 1.13 g/L | 1.13 g |
| diH2O | – | 900 mL (fill up to) |
| 20% glucose (optional, depending on storage) | 2% | 100 mL |
| Total | n/a | 1,000 mL |
Prepare 1 L of SCD liquid media by adding all components in the order described in the table to 500 mL of diH2O. Allow each component to fully dissolve before adding the next one by stirring on a stir plate with a stir bar. Remove the stir bar and bring the volume to 900 mL. Split the liquid evenly between two 1 L flasks and autoclave (see Table 1). Store the autoclaved medium at room temperature or 4 °C. Add 100 mL of the filter-sterilized glucose solution, which brings the glucose concentration to 2% and the volume to 1 L. Store at 4 °C.
7. YPD liquid media
| Reagent | Final concentration | Quantity or volume (for 1 L) |
|---|---|---|
| BactoTM yeast extract | 10 g/L | 10 g |
| BactoTM peptone | 20 g/L | 20 g |
| diH2O | – | 900 mL (fill up to) |
| 20% glucose | 2% | 100 mL |
| Total | n/a | 1,000 mL |
Prepare 1 L of YPD liquid media by adding all components in the order described to 500 mL of diH2O. Allow each component to fully dissolve before adding the next one by stirring on a stir plate with a stir bar. Remove the stir bar and bring the flask to 900 mL. Split the liquid evenly between two 1 L flasks and autoclave for 30 min using the liquid autoclaving cycle (Table 1). When the medium cools down to 40–50 °C, add 100 mL of the filter-sterilized glucose solution and carefully shake, which brings the glucose concentration to 2% and the volume to 1 L. Store at RT.
Critical: The glucose solution or media containing glucose should not be autoclaved to prevent caramelization.
8. YPD agar plates
| Reagent | Final concentration | Quantity or volume (for 1 L) |
|---|---|---|
| BactoTM yeast extract | 10 g/L | 10 g |
| BactoTM peptone | 20 g/L | 20 g |
| Agar, powdered bacteriological grade | 20 g/L | 20 g |
| diH2O | – | 900 mL (fill up to) |
| 20% glucose | 2% | 100 mL |
| Total | n/a | 1,000 mL |
Prepare 1 L of YPD liquid media by adding all components in the order described in the table to 500 mL of diH2O. Allow each component to fully dissolve before adding the next one by stirring on a stir plate with a stir bar. Remove the stir bar and bring the flask to 900 mL. Split the liquid evenly between two 1 L flasks and autoclave for 30 min using the liquid autoclaving cycle (see Table 1). When the medium cools down to 40–50 °C, add 100 mL of the filter-sterilized glucose solution and carefully shake, which brings the glucose concentration to 2% and the volume to 1 L. Pour the hot media into sterile Petri dishes (25 mL per plate) using sterile technique.
Caution: Use insulated gloves to handle the hot flasks when pouring the plates. Allow the plates to cool and dry overnight and store at 4 °C.
9. Life cycle sporulation liquid media (LC-SPO)
| Reagent | Final concentration | Quantity or volume (for 1 L) |
|---|---|---|
| Potassium acetate | 6 g/L | 6 g |
| Sorbitol (from 2% w/v stock solution in diH2O) | 0.02% | 20 mL |
| Adenine hydrochloride hemihydrate | 40 mg/L | 40 mg |
| Uracil | 40 mg/L | 40 mg |
| L-Histidine | 20 mg/L | 20 mg |
| L-Leucine | 20 mg/L | 20 mg |
| L-Tryptophan | 20 mg/L | 20 mg |
| diH2O | – | 1,000 mL (fill up to) |
| 0.25 M Na2CO3 solution (add until pH reaches 8.5) | – | – |
| Total | n/a | 1,000 mL |
Prepare 1 L of LC-SPO liquid media by adding all components in the order described in the table to 500 mL of diH2O. Add the sorbitol volume from a Nalgene-filter-sterilized 2% w/v sorbitol stock solution. Allow each component to fully dissolve before adding the next one by stirring on a stir plate with a stir bar. Remove the stir bar and bring the flask to 1,000 mL. Split the liquid evenly between two 1 L flasks and autoclave for 30 min using the liquid autoclaving cycle (see Table 1). Right before the microfluidic experiment starts, take a 20 mL aliquot of the LC-SPO media in a sterile beaker with a small stir bar and adjust its pH to 8.5 by dropwise adding 0.25 M Na2CO3 solution while constantly measuring the pH and stirring on a stir plate. Make sure the pH meter electrode is well rinsed with diH2O from the potassium chloride storage solution before measuring the pH of the medium. Use the medium immediately; do not store. This medium is a modification of the sporulation medium from Kociemba et al. [16].
10. Stock Nystatin solution in DMSO
Prepare 3 mg/mL of Nystatin by adding 30 mg of Nystatin powder to 5 mL of DMSO. Fill up to 10 mL. Once all Nystatin has dissolved, dispense 1 mL aliquots into 1.7 microtubes and store at -20 °C.
Laboratory supplies
1. Petri dishes (KORD-Valmark, catalog number: 2900)
2. 15 mL Falcon tubes, bulk (Olympus plastics, catalog number: 28-103)
3. 1.7 mL microtube (Eppendorf tubes), sterile (Olympus plastics, catalog number: 22-284)
4. Inoculation loops 10 μL, flexible, PP 20/peel bag (FisherbrandTM Disposable Inoculating Loops, catalog number: 22-363-600 or similar)
5. Ergonomic pipette tips for research, 10 μL (Olympus Plastics, catalog number: 24-130RS)
6. Microfluidic plate for haploid yeast cells (4 chamber, 3.5–5 μm) (CellASIC® ONIX2, catalog number: Y04C-02-5PK)
7. Nalgene® bottle-top sterile, capacity 500 mL, pore size 0.2 μm, fits 45 mm bottle neck (Sigma Millipore, catalog number: Z358223)
8. 2 mL external threaded polypropylene cryogenic vial, self-standing with round bottom (Corning, catalog number: 430659)
9. Electrode storage solution, potassium chloride pH kit (OHAUS, catalog number: 01-922-060)
10. Carl Zeiss immersion oil (refractive index = 1.518, fluorescence free, ISO 8036-1/2 certified, catalog number: NC2187819)
11. Creativity Street Modeling Clay 220g-Neon Colors (Creativity Street, a Dixon Ticonderoga Brand formerly known as Pacon, catalog number: PAC4091, UPC # 021196040915)
12. Olympus spectrophotometry cuvettes, semi-micro (Genesee, catalog number: 21-136)
13. KIMTECH SCIENCE® KIMWIPESTM Delicate Task Wipers (KIMBERLY-CLARK PROFESSIONAL, catalog number: 89218-057)
Equipment
1. Incubator myTemp (Benchmark, model: H2265HC)
2. Nanodrop (Thermo Scientific, model: NanoDrop ONEC)
3. Incu-shaker Mini (Benchmark, model: H1001-M)
4. Vortex mixer (Benchmark, model: BV1000)
5. Ultrasonic homogenizer (Benchmark, model: PULSE 150)
6. Benchtop phase microscope (Zeiss, model: Primostar 3)
7. Mini-centrifuge (PrismTM Mini Centrifuge, catalog number: Z763098); this item has been discontinued, but other mini centrifuges will work, e.g., OHAUSTM FrontierTM 5306 Mini Centrifuge, catalog number: S43248
8. Zeiss Observer Z1 microscope controlled by ZEN pro software with the following specs: a motorized stage, temperature control for 25 °C, a 40× Zeiss EC Plan-Neofluar 40× 1.3 NA oil Ph 3 M27 immersion objective, the focusing system Definite Focus 3.0, an AxioCam 712 monochrome camera, and a X-CITE XYLIS XT720S lamp (Excelitas Technologies) as light source; however, any microscope with a phase contrast objective and a multiposition focusing system can be used
9. AquasearcherTM pH meter (OHAUS, model: a-AB23PH)
10. pH electrode (OHAUS, model: ST320PH/ATC)
11. CellASIC® ONIX2 Microfluidic System (EMD Millipore Corporation, Merck, model: CAX2-S0000)
Software and datasets
1. Open-Source software 1: Yeastvision, a pip-installable open-source GUI-based framework for deep-learning-enabled segmentation, tracking, and time-series analysis of the full Saccharomyces cerevisiae life cycle. Version 0.1.67. https://pypi.org/project/yeastvision/
2. Imaging data showing the execution of the protocol have been deposited to Dryad (Dataset S1): https://doi.org/10.5061/dryad.3bk3j9kw0
Procedure
文章信息
稿件历史记录
提交日期: Aug 26, 2025
接收日期: Oct 27, 2025
在线发布日期: Nov 18, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kennedy, T., Neupane, S., Merritt, B. and Argüello-Miranda, O. (2025). Imaging the Entire Sexual Life Cycle of the Budding Yeast Saccharomyces cerevisiae Using a Microfluidic Platform. Bio-protocol 15(23): e5536. DOI: 10.21769/BioProtoc.5536.
- Kennedy, T., Yalcinkaya, B., Ramakanth, S., Neupane, S., Tadic, N., Buchler, N. E. and Argüello-Miranda, O. (2025). Deep learning-driven imaging of cell division and cell growth across an entire eukaryotic life cycle. Mol Biol Cell. mbcE25010009. https://doi.org/10.1091/mbc.E25-01-0009
分类
微生物学 > 微生物细胞生物学 > 细胞成像
细胞生物学 > 细胞成像 > 微流体
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link