- EN - English
- CN - 中文
An Optimized Protocol for High-Quality AFM Imaging of Amyloid Fibrils
获得高质量淀粉样纤维 AFM 成像的优化实验流程
发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5533 浏览次数: 1288
评审: Elena A. OstrakhovitchSanjay Kumar KureelAnonymous reviewer(s)
Abstract
Characterizing the morphology of amyloid proteins is an integral part of studying neurodegenerative diseases. Such morphological characterization can be performed using atomic force microscopy (AFM), which provides high-resolution images of the amyloid protein fibrils. AFM is widely employed for visualizing mechanical and physical properties of amyloid fibrils, not only from a biological and medical perspective but also in relation to their nanotechnological applications. A crucial step in AFM imaging is coating the protein of interest onto a substrate such as mica. However, existing protocols for this process vary considerably. The conventional sample preparation method often introduces artifacts, particularly due to deposition of excess salt. Hence, an optimized protocol is essential to minimize salt aggregation on the mica surface. Here, we present an optimized protocol for coating amyloid proteins onto mica using the dip-washing method to eliminate background noise. This approach improves the adherence of protein to the mica surface while effectively removing residual salts.
Key features
• The protocol introduces a new method to coat protein samples onto mica sheets for AFM imaging.
• It presents a dip-washing technique aimed at removing excess salt deposited on the mica sheet, thereby minimizing imaging artifacts.
• This protocol can be used for analyzing amyloid fibrillation mechanisms as well as capturing time-dependent fibrillation dynamics under various physiological conditions.
• It also provides clear stepwise washing instructions that balance the salt removal and retention of protein fibrils on the mica.
Keywords: Amyloid fibril (淀粉样纤维)Graphical overview
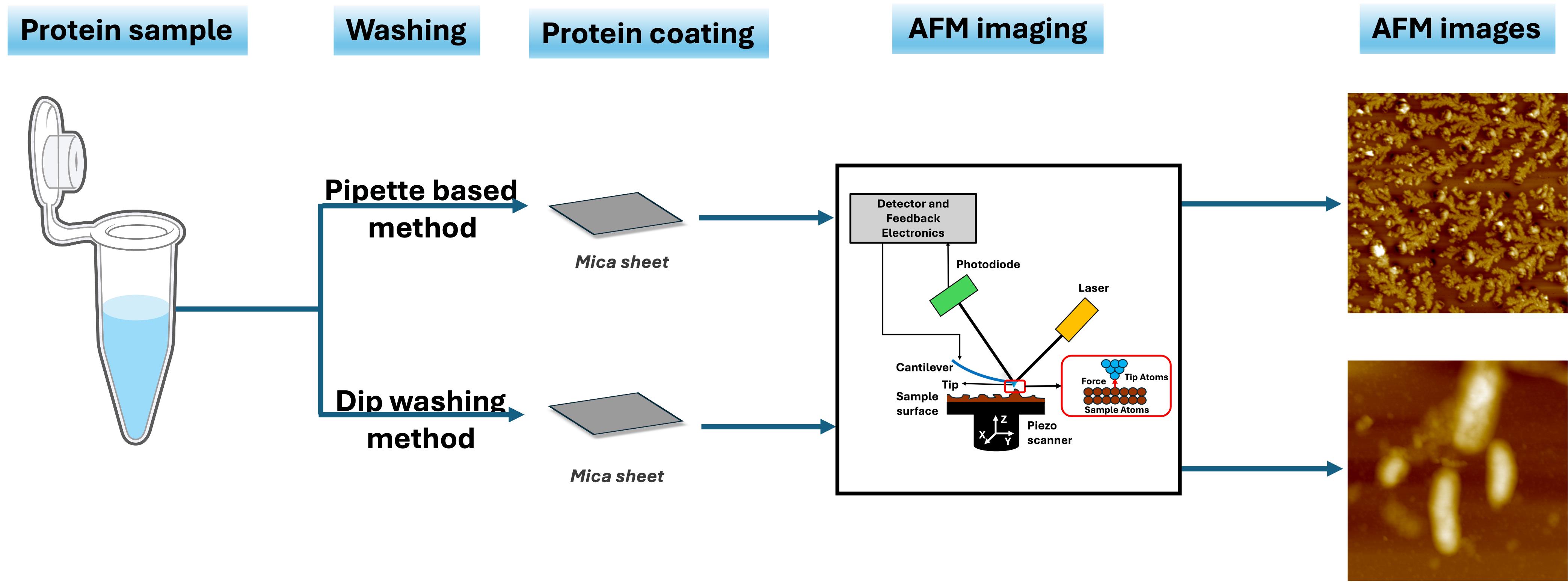
Background
Amyloids, which are associated with numerous neurodegenerative diseases like Alzheimer's and Parkinson's, are characterized by their tendency to form insoluble fibrillar aggregates that impact neuronal functions by triggering neuroinflammation [1–3]. Gaining insights into the structural features of their fibril morphology is critical for elucidating not only the pathological mechanisms but also therapeutic development [4]. Atomic force microscopy (AFM) is a powerful technique for imaging amyloid fibril morphologies, utilizing advanced techniques such as tapping-mode and high-speed AFM [5–7]. Notably, studies have shown that tapping-mode AFM, which is frequently used, does not compromise fibrillar integrity because it minimizes the interaction between the tip and sample [8–11]. Several investigations have been conducted under different environmental conditions, ranging from low to extreme salt concentrations, to study their influence on amyloid morphologies [12–18]. However, caution must be taken during sample preparation, as it is a crucial aspect of AFM imaging. Improper or inadequate washing can lead to salt artifacts or protein loss, which may compromise image quality and affect data interpretation. For instance, the presence of excess salt in the sample can cause obstacles in AFM, thereby reducing the accuracy of amyloid fibril imaging [19,20]. Therefore, a standardized washing protocol must be followed in a manner that prevents salt artifacts while minimizing amyloid loss to maintain image quality [21]. The commonly used approach of pipette-based coating of amyloid fibrils onto mica sheets, followed by pipette-based rinsing and subsequent drying, may result in inconsistent fibril adsorption, limited reproducibility, and introduction of aggregation-related artifacts [22]. This realization has recently led to the development of a software tool based on a deep learning method to denoise salt artifacts from AFM images of amyloid proteins [23].
Materials and reagents
Reagents
1. Autoclaved Milli-Q water
2. APTES [(3-aminopropyl) triethoxysilane] (Sigma-Aldrich, catalog number: 440140)
3. Sodium phosphate monobasic monohydrate (Amresco, catalog number: 0823)
4. di-sodium hydrogen orthophosphate (Qualigen, catalog number: Q15855)
5. Sodium chloride (SR Life Science, catalog number: 41721)
Solutions
1. Phosphate buffer with low salt concentration (10 mM NaCl) (see Recipes)
2. Phosphate buffer with physiological salt concentration (150 mM NaCl) (see Recipes)
Recipes
1. Phosphate buffer with low salt concentration (10 mM NaCl)
10 mM sodium phosphate (pH 8.0) (sodium phosphate monobasic monohydrate and di-sodium hydrogen orthophosphate)
10 mM sodium chloride
2. Phosphate buffer with physiological salt concentration (150 mM NaCl)
10 mM sodium phosphate (pH 8.0)
150 mM sodium chloride
Laboratory supplies
1. Mica sheets grade V-1, 15 × 15 × 0.15 mm (SPI Chem, catalog number: 01868-CA)
2. 50 mL volume beaker (Figure 1a)
3. Kimwipes disposable wipers (Kimberly Clark Hygiene Products Pvt Ltd., catalog number: 34155)
4. 1.5 mL volume microcentrifuge tubes (Tarsons, catalog number: 500016) (Figure 1a)
5. Tube holders or stands (Figure 1a)
6. Round Petri plates (Tarsons, catalog number: 460030) (Figure 1a)
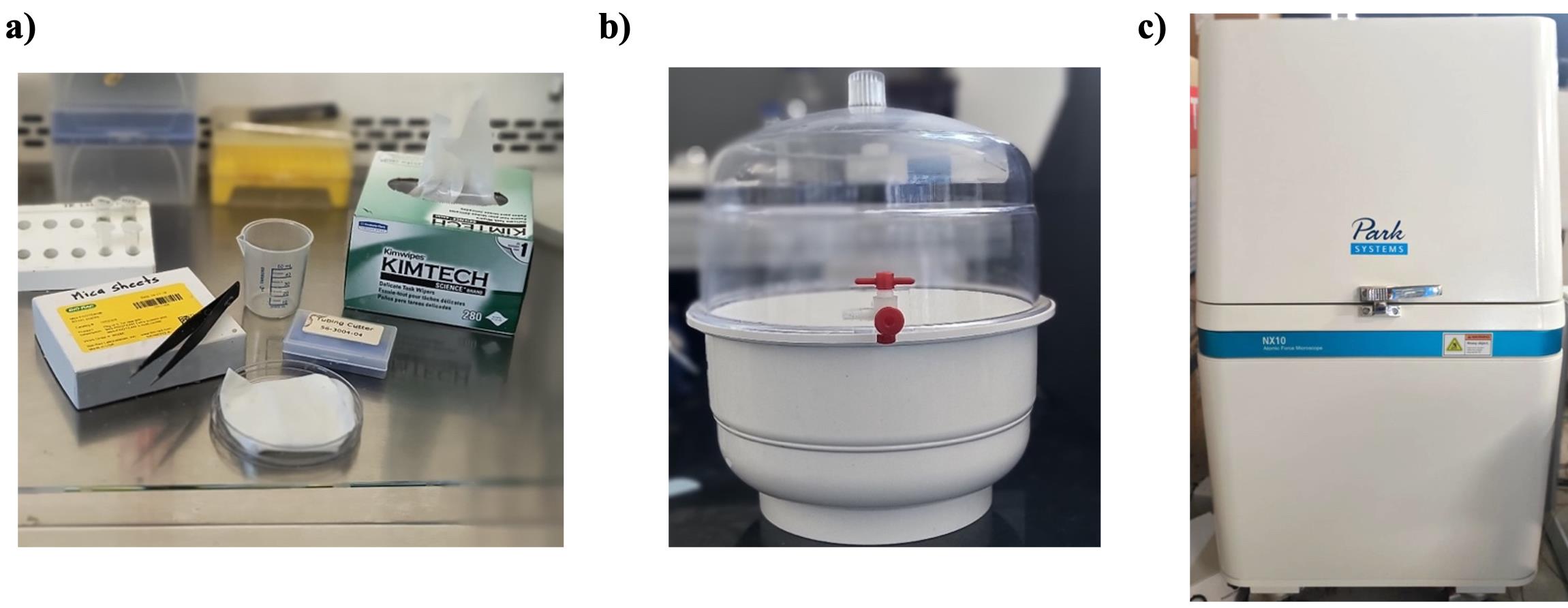
Figure 1. Materials used in this protocol. (a) Experimental setup. (b) Desiccator. (c) Park Systems NX10 AFM instrument.
Equipment
1. Mica sheet cutter: sharp blade or scissors
2. Forceps
3. Desiccator (Figure 1b)
4. Park Systems NX10 atomic force microscope (Figure 1c)
5. Cantilever (Park Systems, catalog number: NSC36_C)
Software and datasets
1. XEI software (for AFM data acquisition and processing)
Procedure
文章信息
稿件历史记录
提交日期: Sep 9, 2025
接收日期: Nov 2, 2025
在线发布日期: Nov 13, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Kumar, R. R., Das, U., Uttamrao, P. P. and Rathinavelan, T. (2025). An Optimized Protocol for High-Quality AFM Imaging of Amyloid Fibrils. Bio-protocol 15(23): e5533. DOI: 10.21769/BioProtoc.5533.
分类
生物物理学 > 显微技术 > 原子力显微镜
生物化学 > 蛋白质 > 成像
神经科学 > 神经系统疾病 > 神经退行性病变
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











