- EN - English
- CN - 中文
Bridging PCR-Based Genome-Walking Protocol
基于 Bridging PCR 的基因步移实验流程
发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5531 浏览次数: 1035
评审: Anu ThomasAlberto RissoneAnonymous reviewer(s)

相关实验方案

一种快速且经济的流程,用于从细菌草图基因组中识别并捕获生物合成基因簇
Marco A. Campos-Magaña [...] Luis Garcia-Morales
2025年12月20日 778 阅读
Abstract
Genome walking is a classical molecular biology technique used to amplify unknown regions flanking known DNA sequences. Genome walking holds a vital position in the areas associated with molecular biology. However, existing genome-walking protocols still face issues in experimental operation or methodological specificity. Here, we propose a novel genome-walking protocol based on bridging PCR. The critical factor of this protocol is the use of a bridging primer, which is made by attaching an oligomer (or tail primer sequence) to the 5′ end of the walker primer 5′ region. When the bridging primer anneals to the walker primer site, this site will elongate along the tail of the bridging primer. The non-target product (the main contributor to background in genome walking), defined by the walker primer, is lengthened at both ends. In the next PCR(s), the annealing between the two lengthened ends is easier than the annealing between them and the shorter tail primer. As a result, this non-target product is eliminated without affecting target amplification.
Key features
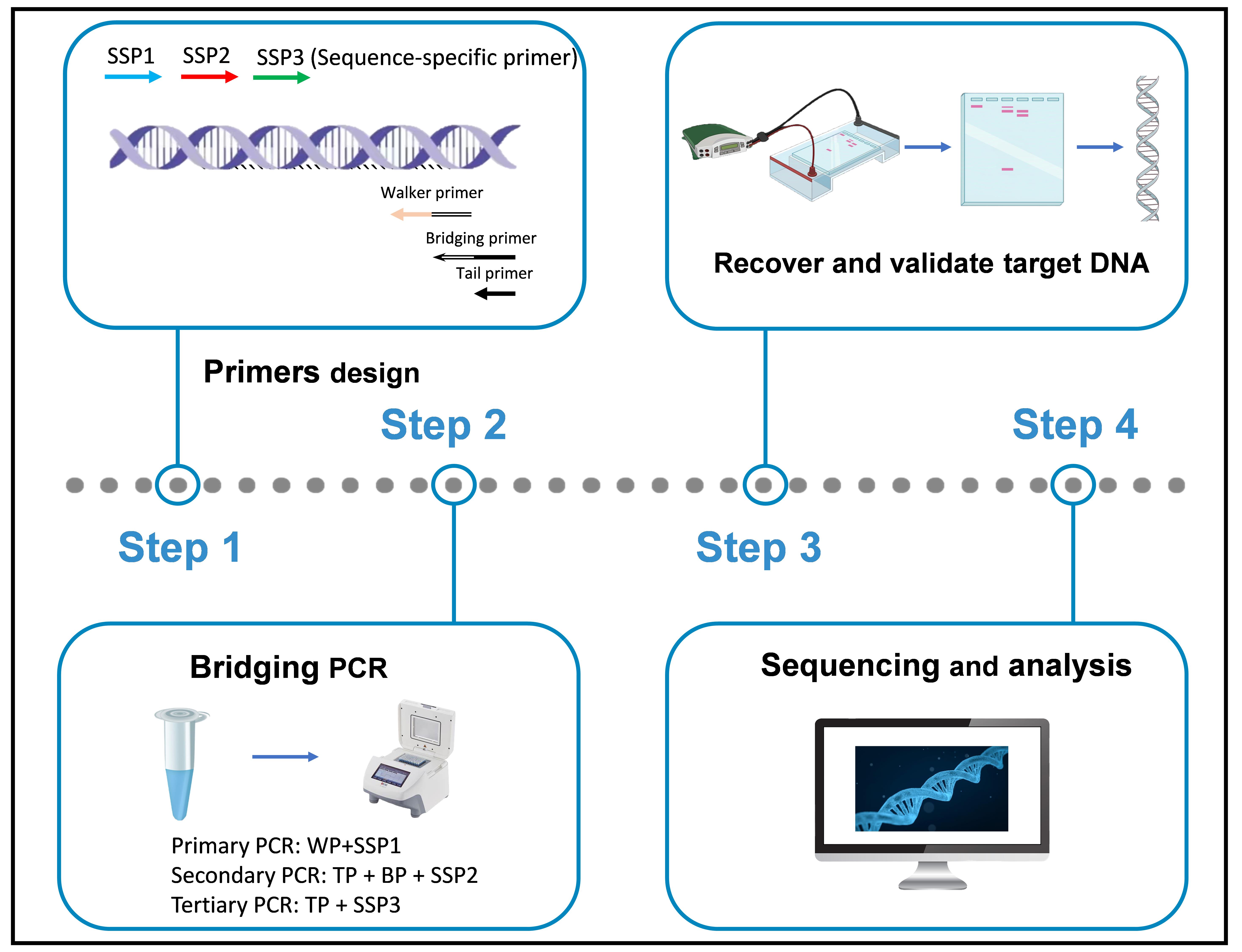
• This bridging PCR protocol, built upon the technique developed by Lin et al. [1], is universal.
• The bridging primer is made by attaching a tail DNA to the 5′ end of the walker primer 5′ region.
• Lengthening of non-target DNA by both ends of bridging primer results in intrastrand annealing or hairpin formation, the basis for the removal of non-target background.
Keywords: Bridging PCR (Bridging PCR)Graphical overview
Background
Genome walking (GW) is a molecular tool for cloning unknown regions flanking known DNAs, facilitating methods such as gene cloning, identifying DNA mutations, and analyzing transgenic sites [2–5]. Up to date, three types of GW were available: random PCRs, genome library-based techniques, and restriction-ligation-based PCRs. Among them, genome library-based techniques are time-consuming and inefficient, due to requiring the construction and screening of a genomic DNA library, and restriction-ligation-based PCRs require the digestion of genomic DNA and the subsequent ligation of the digested product prior to amplification. Comparatively, random PCRs are faster and more efficient, as the extra steps prior to amplification are omitted [6–10].
Random PCRs generally require two to three rounds of nested amplification. In primary amplification, a low-temperature annealing cycle allows the walking primer (WP) to randomly anneal to the unknown flank, thereby synthesizing a target DNA comprising a known region and its unknown flank. The following round or two of nested amplification further enrich this DNA, ultimately achieving the so-called genome walking [11–15]. However, due to the use of WP and at least one low-temperature cycle in each round of amplification, three types of non-target amplicons will be produced. Type I is synthesized by GSP alone; type II is synthesized by GSP and WP; and type III is synthesized by WP alone. Types I and II can be easily removed in the next PCR because they lack an authentic binding site for the sequence-specific primer (SSP). The real challenge is how to eliminate the type III non-target product [16–19]. Existing random PCRs, such as thermal asymmetric interlaced PCR [20], fusion primer and nested integrated PCR [21], and partially overlapping primer-based PCR [22,23], have their reliability compromised due to the accumulation of this non-target product. Therefore, a truly reliable genome-walking scheme should be able to fundamentally overcome this non-target amplification, which has always been pursued by researchers [24–27].
In this study, a bridging PCR-based genome-walking protocol was designed. The main innovation of this PCR is the use of a bridging primer (BP) in secondary PCR, which is made by attaching an oligomer (or tail primer, TP) to the 5′ end of the WP 5′ region. As a result, in secondary PCR, the primary non-target product defined by the WP—namely, the main contributor to background—is lengthened by the BP at both ends. Clearly, this DNA itself preferentially forms a hairpin via intrastrand annealing between the lengthened ends, instead of being amplified by the TP. In contrast, the amplification of the primary target DNA is not affected, because it is defined by both the SSP and WP. The feasibility of the bridging PCR has been validated by extending into unknown flanking regions of several known genes. Overall, this bridging PCR could be an alternative to existing genome-walking methods.
Materials and reagents
Biological materials
1. Genome of Levilactobacillus brevis CD0817 [28–33], extracted using the Bacterial Genomic DNA Isolation kit (Tiangen Biotech Co., Ltd., Beijing, China)
Reagents
1. 10× LA PCR buffer (Mg2+ plus) (Takara, catalog number: RR042A)
2. 6× Loading buffer (Takara, catalog number: 9156)
3. LA Taq polymerase (hot-start version) (Takara, catalog number: RR042A)
4. dNTP mixture (Takara, catalog number: RR042A)
5. DL 5,000 DNA marker (Takara, catalog number: 3428Q)
6. 1× TE buffer (Sangon, catalog number: B548106)
7. Agarose (Sangon, catalog number: A620014)
8. 1 M NaOH (Yuanye, catalog number: B28412)
9. 0.5 M EDTA (Solarbio, catalog number: B540625)
10. Goldview nucleic acid gel stain (10,000×) (Yeasen, catalog number: 10201ES03)
11. Tris (Solarbio, catalog number: T8060)
12. Boric acid (Solarbio, catalog number: B8110)
13. TaKaRa MiniBEST DNA Fragment Purification kit v4.0 (TaKaRa, catalog number: DV9761)
14. Primers (Sangon)
WP1: GTCGTAGTCATGTATCGTCCTAGTCATCTGCTTGTTCGTCAGTCAGCGTC
WP2: GTCGTAGTCATGTATCGTCCTAGTCTCAGTCAGTCAGTTGCAGTCAGTCT
WP3: GTCGTAGTCATGTATCGTCCTAGTCATCCAGAACAGTCGATTGGTTCAGC
BP: CAGTCAGTCTCAGCTAGTCAGTGTCGTCGTAGTCATGTATCGTCCTAGTC
TP: CAGTCAGTCTCAGCTAGTCAGTGTC
gadA-SSP1: TCCAAGAATCATCCGCAATCGTCA
gadA-SSP2: TGGTAACATCGTCACGGTTCTTTGG
gadA-SSP3: TAGCCTTGTACCCATCTTTACCGAA
gadR-SSP1: TCCTTCGTTCTTGATTCCATACCCT
gadR-SSP2: CCATTTCCATAGGTTGCTCCAAGG
gadR-SSP3: GGATACTGGCTAAAATGAATTAACTCGGATAA
hyg-SSP1: ACGGCAATTTCGATGATGCAGCTTG
hyg-SSP2: GGGACTGTCGGGCGTACACAA
hyg-SSP3: CTGGACCGATGGCTGTGTAGAAG
Solutions
1. 2.5× TBE buffer (see Recipes)
2. 0.5× TBE buffer (see Recipes)
3. 100 mM primer (see Recipes)
4. 10 mM primer (see Recipes)
5. 1.5% agarose gel (see Recipes)
Recipes
1. 2.5× TBE buffer (pH 8.3)
| Reagent | Final concentration | Amount |
|---|---|---|
| 0.5 M EDTA solution | 5 mM | 10 mL |
| Tris | 225 mM | 27 g |
| Boric acid | 225 mM | 13.75 g |
| Ultrapure water | n/a | n/a |
| Total | n/a | 1,000 mL |
This 2.5× TBE buffer can be stored at room temperature for 3 months.
2. 0.5× TBE buffer (pH 8.3)
| Reagent | Final concentration | Amount |
|---|---|---|
| 2.5× TBE buffer | 0.5× | 200 mL |
| Ultrapure water | n/a | 800 mL |
| Total | n/a | 1,000 mL |
This 0.5× TBE buffer can be stored at room temperature for 3 months.
3. 100 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Primer powder | 100 μM | n/a |
| 1× TE buffer | 1× | Volume specified by the supplier |
| Total | n/a | Volume specified by the supplier |
Note: Take 10 μL of this primer solution to make 10 μM primer, and store the remaining solution at -80 °C.
4. 10 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 100 μM primer | 10 μM | 10 μL |
| 1× TE buffer | 1× | 90 μL |
| Total | n/a | 100 μL |
Note: Divide 10 μM primer into 10 μL/tube, then store these tubes at -20 °C.
5. 1.5% agarose gel
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1.5% | 1.5 g |
| 0.5× TBE buffer | 0.5× | 100 mL |
| Goldview nucleic acid gel stain (10,000×) | 1× | 10 μL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 0.2 mL PCR tubes (Kirgen, catalog number: KG2311)
2. 10 μL pipette tips (Sangon, catalog number: F600215)
3. 200 μL pipette tips (Sangon, catalog number: F600227)
4. 1,000 μL pipette tips (Sangon, catalog number: F630101)
5. 1,500 μL microcentrifuge tubes (Labselect, catalog number: MCT-001-150)
Equipment
1. PCR apparatus (Applied Biosystems, model: Biometra TAdvanced 96 PCR)
2. Electrophoresis apparatus (Beijing Liuyi, model: DYY-6C)
3. Gel imaging system (Bio-Rad, model: ChemiDoc XRS+)
4. Microcentrifuge (Tiangen, model: TGear)
Software and datasets
1. Oligo v7.37 software (Molecular Biology Insights, Inc., USA)
2. DNASTAR Lasergene v7.1 software (DNASTAR, Inc., USA)
Procedure
文章信息
稿件历史记录
提交日期: Sep 22, 2025
接收日期: Nov 2, 2025
在线发布日期: Nov 11, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Li, M., Gu, Y., Tang, Q. and Li, H. (2025). Bridging PCR-Based Genome-Walking Protocol. Bio-protocol 15(23): e5531. DOI: 10.21769/BioProtoc.5531.
分类
分子生物学 > DNA > 基因组步移
分子生物学 > DNA > PCR
分子生物学 > DNA > DNA 克隆
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link