- EN - English
- CN - 中文
Utilizing EdU to Track Leukocyte Recruitment to the Brain
使用 EdU 追踪脑部白细胞募集
发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5527 浏览次数: 1564
评审: Ivonne SehringAnonymous reviewer(s)
Abstract
Detecting the proliferation of cells with copper(I)-catalyzed azide-alkyne cycloaddition (click chemistry) and the thymidine analogue, 5-ethynyl-2’-deoxyuridine (EdU), is a simpler and more versatile method than traditional antibody-based approaches. Instead of the harsh series of steps typically used for 5-bromo-2’-deoxyuridine (BrdU) detection, detecting EdU does not require DNA denaturation and is suitable for use with other applications. This approach was implemented in an animal model of ischemic stroke. The following protocol details how to use EdU to label, track, and visualize leukocyte recruitment for flow cytometry and fluorescence microscopy, including the processes for EdU injection and blood and tissue sample preparation. Considerations for timing, dosing, and cell viability are also outlined to tailor the protocol to experimental needs. This method could be applied to various models that require extended tracking periods, as the signal from EdU can last several cell divisions, depending on cell type and condition.
Key features
• EdU labeling is simple and compatible with routine laboratory methods.
• This method is compatible with genetically encoded fluorophores, such as GFP and tdTomato.
• EdU incorporation in circulating leukocytes varies depending on the specific cell subtype.
• This technique can be adapted to track leukocyte recruitment, including when cells are recruited from the bloodstream.
Keywords: EdU (EdU)Graphical overview
Background
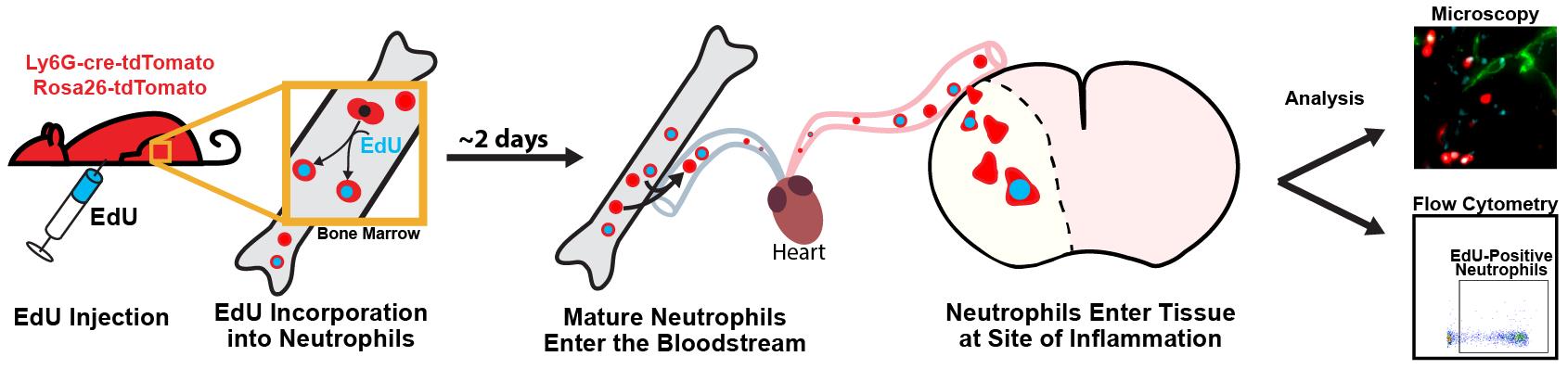
Leukocytes play a key role in the body’s inflammatory response, and understanding the patterns and activity of leukocyte recruitment yields valuable insight for therapeutic interventions. Cell-tracking tools like the nucleotide analogues 5-ethynyl-2’-deoxyuridine (EdU) and 5-bromo-2’-deoxyuridine (BrdU) help to reveal the behavior of leukocytes, as well as many other cell types, across a wide range of pathological processes [1,2]. With EdU labeling, cells incorporate EdU into their DNA during replication, mimicking the nucleoside thymidine. A copper(I)-catalyzed reaction then allows for a fluorescent azide to attach to the alkyne group (click chemistry) on EdU, enabling the detection of dividing cells [2,3]. This is similar to BrdU; however, EdU has greater utility for multiplexed staining and minimizes the disruption of protein epitopes because it does not require DNA denaturation for visualization [4,5]. EdU can be injected intraperitoneally for rapid labeling or given through food or drinking water to prolong the labeling window. EdU is incorporated into the DNA of cells during the S phase of the cell division cycle. As such, for myeloid cells like neutrophils and monocytes, it is incorporated into immature precursors in the bone marrow, which enter the bloodstream after maturation [6–8]. EdU injections could be staggered over several hours or days to label a higher proportion of cells, if desired [9]. One current limitation with EdU compared to other cell-labeling protocols, such as carboxyfluorescein succinimidyl ester or fluorescent antibodies, is that EdU cannot be visualized for live-cell imaging, since the sample must be fixed to detect the EdU signal. Additionally, high concentrations of EdU or prolonged exposure could have cytotoxic effects and may impact cell viability [10].
Here, we detail a protocol for using EdU to label and track leukocytes that also have a genetically encoded fluorophore in an ischemia/reperfusion model of ischemic stroke in mice. While it is optimized for leukocytes, this method can be adapted to investigate other cell types and aims [11,12]. This model demonstrates the dynamics of leukocyte recruitment following an inflammatory event, including which cells are preferentially recruited, how long they circulate in the blood or persist in tissue, and the fraction of each leukocyte subset involved in the response [13]. The methodology here can be used to unobtrusively label a wave of leukocytes in circulation. The labeled leukocytes can be identified in tissue at a later point to gain insights into their location, recruitment patterns, and enrichment.
Materials and reagents
Biological materials
1. Transgenic mice with fluorescent reporter(s)
Note: Here, we describe methods done with the Catchup mouse line, which is on the C57BL/6 background [14], but the methodology could be applied to other mouse lines and reporters. The Catchup mouse has cre-ERT2 and tdTomato inserted into the Ly6G locus and also the Rosa26-CAG-tdTomato reporter. This results in >95% of the circulating neutrophils expressing the red fluorescent protein, tdTomato.
Reagents
1. 2'-Deoxy-5-ethynyluridine (EdU) (Biosynth, catalog number: NE08701)
2. Ammonium-chloride-potassium (ACK) lysing buffer (VWR International, catalog number: 10128-802)
3. Accutase (Gibco, catalog number: A11105)
4. Alexa Fluor 647 conjugated Azide Plus (picolyl azide) (Vector Labs, catalog number: CCT-1482)
5. Copper (II) sulfate pentahydrate (Sigma-Aldrich, catalog number: 209198)
6. Phosphate-buffered saline (PBS), without calcium and magnesium, 10× concentration (Corning, catalog number: 20-031-CV)
7. PBS (without calcium and magnesium), 1× concentration (Corning, catalog number: 21-031-CV)
8. Ethylenediaminetetraacetic (EDTA) powder (Sigma-Aldrich, catalog number: E9884)
9. DyLight 650 Antibody Labeling kit (Thermo Fisher, catalog number: 84535)
10. Fetal bovine serum (FBS) (GeminiBio, catalog number: 100-500)
11. Glycerol (Sigma-Aldrich, catalog number: 5516)
12. Hank’s balanced salt solution (HBSS) with calcium, magnesium, no phenol red, 10× concentration (Gibco, catalog number: 14065-056)
13. HBSS with calcium, magnesium, and phenol red, 1× concentration (Corning, catalog number: 21-020-CV)
14. Hydrochloric acid (HCl) (37%) (VWR Chemicals, catalog number: 7647-01-0)
15. NP-40 (10%) (Thermo Fisher, catalog number: 28324)
16. Optimal cutting temperature (OCT) compound (Thermo Fisher, catalog number: 23-730-571)
17. Paraformaldehyde (PFA) powder (Sigma-Aldrich, catalog number: 158127)
18. Percoll (Sigma-Aldrich, catalog number: P1644)
19. Sodium ascorbate (Sigma-Aldrich, catalog number: 134-03-2)
20. Sodium chloride (NaCl) (Thermo Fisher, catalog number: BP358-212)
21. Sodium hydroxide (NaOH) (Thermo Fisher, catalog number: S320-500)
22. Sucrose (Thermo Fisher, catalog number: BP220-1)
23. Tris base, white crystals (Thermo Fisher, catalog number: BP152-1)
24. Triton X-100 (Sigma-Aldrich, catalog number: 9002-93-1)
25. 2,3,5-Triphenyltetrazolium chloride (Sigma-Aldrich, catalog number: T8877)
26. Zombie violet live dead stain (BioLegend, catalog number: 423114)
Antibodies
1. Anti-Fc receptor (BD Biosciences, catalog number: 553142, stock concentration 500 μg/mL)
2. Anti-CD45 antibody-BV510 conjugated (BioLegend, catalog number: 103138, stock concentration 200 μg/mL)
3. Anti-CD11b antibody-BV650 conjugated (BioLegend, catalog number: 101259, stock concentration 200 μg/mL)
4. Anti-PECAM antibody, clone 390 (Millipore-Sigma, catalog number: CBL1337, stock concentration 500 μg/mL)
Solutions
1. EdU solution (see Recipes)
2. EDTA solution (see Recipes)
3. 40% Percoll solution (see Recipes)
4. Fluorescence-activated cell sorting (FACS) buffer (see Recipes)
5. 4% Formaldehyde solution (see Recipes)
6. Permeabilization buffer (see Recipes)
7. Fluorophore-conjugated Azide Plus (see Recipes)
8. Copper (II) sulfate pentahydrate solution (see Recipes)
9. Sodium ascorbate solution (see Recipes)
10. 5 N sodium hydroxide (see Recipes)
11. Tris-buffered saline (TBS buffer) (see Recipes)
12. 2% Triphenyltetrazolium chloride (TTC) (see Recipes)
13. 0.5% NP-40 (see Recipes)
14. Click reaction solution (see Recipes)
15. Vessel labeling antibody (see Recipes)
16. 15% sucrose (see Recipes)
17. 30% sucrose (see Recipes)
Recipes
1. EdU solution
For a 5 mg/mL stock, dissolve 50 mg of EdU in 10 mL of 1× PBS.
Note: EdU stock solution should be kept at -20 °C.
2. EDTA solution
For 100 mM EDTA, add 2.92 g of EDTA powder to 80 mL of deionized (DI) H2O while stirring. Add 1.5 g of NaOH and allow for EDTA to begin dissolving. Adjust to final pH 8.0 (with 37% HCl or 5 N NaOH). Adjust final volume to 100 mL with DI H2O.
3. 40% Percoll solution
Add 5 mL of 10× HBSS and 45 mL of 100% Percoll to make 90% Percoll in 1× HBSS. This solution may be stored at 4 °C for 6 months. For 40% Percoll, add 27.8 mL of 1× HBSS to 22.2 mL of the 90% Percoll solution to make the final concentration 40% Percoll in 1× HBSS.
Note: We make this fresh for each experiment.
4. Fluorescence-activated cell sorting (FACS) buffer
For 5% composition, add 2 mL of FBS to 48 mL of 1× PBS; mix and filter through a 0.2 μm filter.
5. 4% Formaldehyde solution (pH 7–7.5)
For 100 mL of solution, heat 85 mL of DI H2O to 55–60 °C on a heater; add 4 g of paraformaldehyde powder while stirring (use a magnetic stir bar). Add 1–3 drops of NaOH and wait for the powder to go into solution. Chill to room temperature and add 10 mL of 10× PBS. Adjust pH to 7.5 (with 37% HCl or 5 N NaOH) as needed, then adjust volume to a final 100 mL with DI H2O and filter through a 0.2 μm filter.
Note: All reagents should be combined in a fume hood. The solution should turn clear in a few minutes after adding the drops of NaOH. Adding NaOH starts a chain reaction that converts the powder into dissolved formaldehyde gas. Care should be taken to ensure the temperature of the solution stays below 60 °C and that it is chilled promptly after the powder dissolves to limit the release of formaldehyde from the solution.
6. Permeabilization buffer
For a 0.5% Triton X-100 solution, add 500 μL of 10% Triton X-100 to 9.5 mL of 1× PBS.
7. Fluorophore-conjugated Azide Plus
For 1 mM Azide Plus, dissolve 1 mg of the powder in 915 μL of DI H2O. Store 20 μL aliquots at -20 °C.
Note: Azide Plus was designed to improve copper binding for a more efficient and quicker conjugation reaction compared to simple azide. Using it allows the researcher to minimize the length of time the sample is in the click solution, as the reaction mixture will decrease the genetic fluorescent signal with time. Additionally, azides conjugated to different fluorophores are sold and can be chosen based on your needs.
8. Copper (II) sulfate pentahydrate solution
For 200 mM, dissolve 0.5 g of copper (II) sulfate pentahydrate in a final volume of 10 mL of DI H2O.
Note: This solution may be stored for a week at room temperature.
9. Sodium ascorbate solution
For 1 M, dissolve 0.3 g of sodium ascorbate in a final volume of 1.5 mL of DI H2O.
Note: We make this solution fresh daily.
10. 5 N sodium hydroxide
For 100 mL, dissolve 20 g of NaOH pellets in 80 mL of DI H2O in a beaker. When cooled, bring the final volume to 100 mL.
11. Tris-buffered saline (TBS buffer)
For 50 mM Tris base with 150 mM NaCl, dissolve 6.05 g of Tris base and 8.76 g of NaCl in 800 mL of DI H2O; adjust pH to 7.5 (with 37% HCl or 5 N NaOH) and bring the final volume to 1 L.
12. 2% triphenyltetrazolium chloride (TTC)
For 2%, dissolve 2.0 g of TTC in 100 mL of PBS.
13. 0.5% NP-40
Add 0.5 mL of 10% NP-40 to 9.5 mL of FACS buffer.
Note: We make this solution fresh daily.
14. Click reaction solution
For 550 μL of solution: start with 542 μL of TBS buffer and add 1.4 μL of copper (II) sulfate pentahydrate solution, 1.1 μL of 1 mM Azide Plus solution, and 5.5 μL of sodium ascorbate solution. Final concentrations: 50 mM Tris, 150 mM NaCl, 0.5 mM copper sulfate, 2 μM Azide Plus reagent, and 10 mM sodium ascorbate.
Note: The click reaction should be prepared in the order listed and used within 10 min of preparation. Total volume is sufficient to label one sample for flow cytometry or one 1 mm or 2 mm slice in a 24-well dish.
15. Vessel labeling antibody
To label blood vessels, we use anti-PECAM antibody that we conjugate to DyLight 650 in the lab according to the manufacturer’s protocol. The volume of the antibody stock is adjusted to 0.5 mg/mL with 10× PBS, DI H2O, and glycerol so that the final concentration is 1× PBS and 40% glycerol. The stock solution is stored at -20 °C. To label vessels, 40 μL of conjugated antibody stock is diluted into 100 μL of 1× PBS and injected retro-orbitally 30 min before euthanizing the animal.
Note: Several different fluorophores are available for conjugation. We found that far red fluorophores work well when imaging brain tissue, whereas green fluorophores often do not rise above background in brain tissue for widefield imaging, particularly with thicker samples (>0.5 mm). Alternatively, fluorophore-conjugated lectins could be used. We have had good success with tomato lectin.
16. 15% sucrose
For 100 mL of solution, mix 80 mL of DI H2O with 10 mL of 10× PBS and 5.76 g of sucrose. Adjust the pH to 7.5 (with 37% HCl or 5 N NaOH) and bring the final volume to 100 mL.
17. 30% sucrose
For 100 mL of solution, mix 80 mL of DI H2O with 10 mL of 10× PBS and 11.53 g of sucrose. Adjust the pH to 7.5 (with 37% HCl or 5 N NaOH) and bring the final volume to 100 mL.
Laboratory supplies
1. Magnetic stir bar
2. Brain matrix coronal block (World Precision Instruments, catalog number: RBMS-200C)
3. Coverslips (MatTek, catalog number: PCS-1.5-10)
4. Coverslip dish (MatTek, catalog number: P35G-1.5-10-C)
5. 15 mL tubes (Thermo Fisher, catalog number: 05-539-4)
6. 50 mL conical tubes (Thermo Fisher, catalog number: 339652)
7. 2 mL microcentrifuge tubes (Thermo Fisher, catalog number: 02-681-5)
8. Single-edged razor blades (World Precision Instruments, catalog number: BLADES-2)
9. Straight tip forceps (Sigma-Aldrich, catalog number: F4267)
10. Fine tip forceps (Henry Schein, catalog number: 19-3045)
11. Corneal spring scissors (World Precision Instruments, catalog number: WP2120R)
12. 1 mL syringe (Thermo Fisher, catalog number: 14-823-434)
13. 30 mL syringe (Thermo Fisher, catalog number: 14-823-16K)
14. 25 G needle tip (BD Biosciences, catalog number: 305125)
15. 0.2 μm filter (Sartorius, catalog number: 16532)
16. 70 μm cell strainer (Corning, catalog number: 352350)
17. 100 μm cell strainer (Corning, catalog number: CLS431752)
18. Flow cytometry tubes (Thermo Fisher, catalog number: 50-233-5738)
19. 12-well plate (Thermo Fisher, catalog number: 150200)
20. 24-well plate (Thermo Fisher, catalog number: 142275)
21. Dry ice
22. Superfrost Plus slides (Thermo Fisher, catalog number: 22-034979)
23. Tissue-Tek cryomolds (Sakura, catalog number: 4566)
Equipment
1. Centrifuge (Beckman Coulter, model: GS-6R)
2. Flow cytometer (BD Biosciences, model: LSRFortessa)
3. Widefield microscope (Nikon, model: Eclipse Ni-E)
4. UV cryostat (Leica, model: CM18-60)
Software and datasets
1. NIS Elements Advanced Research Imaging Software (Nikon, version 5.41.02)
2. FIJI image processing software (version 2.17.0)
3. FACSDiva software (BD Biosciences, version 9.0)
4. FlowJo (BD Biosciences, version 10.10), license required
Procedure
文章信息
稿件历史记录
提交日期: Sep 9, 2025
接收日期: Oct 30, 2025
在线发布日期: Nov 11, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Lipfert, Z. K., Arias, E., Batra, A. and Sullivan, D. P. (2025). Utilizing EdU to Track Leukocyte Recruitment to the Brain. Bio-protocol 15(23): e5527. DOI: 10.21769/BioProtoc.5527.
分类
免疫学 > 免疫细胞功能 > 嗜中性粒细胞
细胞生物学 > 细胞运动 > 细胞迁移
神经科学 > 神经系统疾病 > 脑卒中
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link