- EN - English
- CN - 中文
Detailed Protocol for Segmentation and Quantification of Overlapping Prospore Membranes using DeMemSeg
基于 DeMemSeg 的重叠孢膜分割与定量分析的详细操作流程
发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5520 浏览次数: 1339
评审: Hemant Kumar PrajapatiAnonymous reviewer(s)
Abstract
Quantitative analysis of biological membrane morphology is essential for understanding fundamental cellular processes such as organelle biogenesis and remodeling. While manual annotation has been the standard for complex structures, it is laborious and subjective, and conventional automated methods often fail to accurately delineate overlapping objects in 2D projected microscopy images. This protocol provides a complete, step-by-step workflow for the quantitative analysis of overlapping prospore membranes (PSMs) in sporulating yeast. The procedure details the synchronous induction of sporulation, acquisition of 3D fluorescence images and their conversion to 2D maximum intensity projections (MIPs), and the generation of a custom-annotated dataset using a semi-automated pipeline. Finally, it outlines the training and application of our mask R-CNN-based model, DeMemSeg, for high-fidelity instance segmentation and the subsequent extraction of morphological parameters. The primary advantage of this protocol is its ability to enable accurate and reproducible segmentation of individual, overlapping membrane structures from widely used 2D MIP images. This framework offers an objective, efficient, and scalable solution for the detailed quantitative analysis of complex membrane morphologies.
Key features
• Provides a mask R-CNN-based pipeline to accurately segment individual, overlapping membrane structures resulting from 2D maximum intensity projections of 3D image stacks.
• Optimized for quantifying the dynamic morphology of yeast prospore membranes (PSMs), a key model system for studying de novo membrane biogenesis.
• Presents a complete workflow from single-cell isolation using a custom CellPose model to detailed manual annotation for creating high-quality training datasets.
• Enables robust quantitative phenotyping by extracting morphological parameters (e.g., length, roundness) to distinguish subtle differences between wild-type and mutant strains.
Keywords: Deep learning (深度学习)Graphical overview
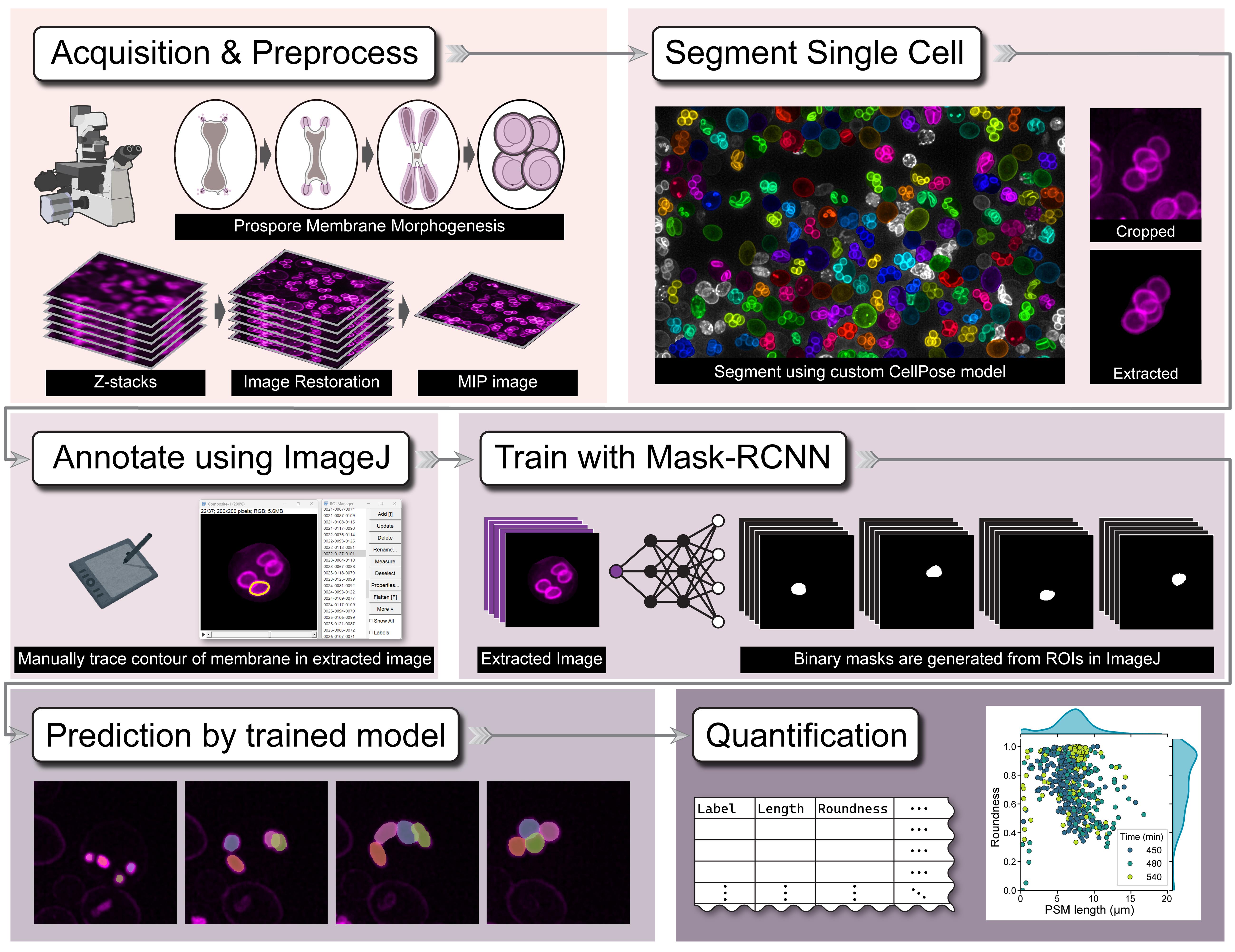
Graphical overview of the DeMemSeg pipeline for automated segmentation and quantitative analysis
Background
Understanding fundamental cellular processes like organelle biogenesis, remodeling, and trafficking often requires quantitative analysis of biological membranes. This involves measuring key morphological features—such as perimeter and roundness—to describe their geometric shape. A key challenge in this field is the accurate segmentation of individual membrane structures from microscopy images, which is often a prerequisite for obtaining reliable quantitative data. The budding yeast prospore membrane (PSM), a model for de novo membrane formation, presents a typical challenge where dynamically growing membranes frequently overlap when visualized in 2D maximum intensity projection (MIP) images.
Historically, segmenting such complex structures has relied on meticulous manual annotation. While often considered the gold standard for accuracy, this method is extremely time-consuming, laborious, and prone to inter-observer variability, making it impractical for large-scale studies. Traditional automated methods based on intensity thresholding [1] or edge detection [2] often fail to accurately delineate boundaries in images with low contrast or, critically, to separate overlapping instances. The advent of generalist deep learning tools, most notably CellPose [3] and ilastik [4], has revolutionized biological image analysis by enabling robust, automated segmentation of individual cells, thereby greatly facilitating single-cell image analysis from dense populations. However, while these tools are highly effective for delineating whole-cell boundaries, their utility is often limited when analyzing more detailed and complex intracellular structures. Specifically, they frequently struggle to resolve extensively overlapping subcellular components like the PSMs within a single cell. Similarly, while large-scale foundation models like the Segment Anything Model (SAM 2) [5] show impressive zero-shot capabilities, our findings indicate they may not achieve the instance-level precision required for resolving the specific, artificial overlaps created by MIP imaging without specialized prompting or fine-tuning.
The DeMemSeg protocol presented here offers several advantages over these existing methods for this specific task. Its primary strength lies in the custom-trained mask R-CNN model, which is specifically optimized to recognize and separate overlapping PSMs in 2D MIP images with high fidelity, a capability where generalist models may falter. By providing a complete, automated workflow from single-cell isolation to instance segmentation, it dramatically improves efficiency and reproducibility compared to purely manual methods. The protocol's utility is demonstrated by its ability to yield quantitative data statistically comparable to manual annotation and to successfully characterize the complex morphological phenotype of an untrained mutant strain. The main limitation of this protocol is its supervised nature, requiring the upfront, labor-intensive creation of a high-quality annotated dataset. Additionally, as a 2D-based analysis, it cannot resolve the true 3D spatial relationships between structures, a general limitation of MIP image analysis.
Beyond yeast PSMs, the principles of this protocol are broadly applicable. The workflow of using a general cell segmentation model (like CellPose) for preprocessing and isolation, followed by training a specialized instance segmentation model (like mask R-CNN) on cropped single-cell images, can be adapted to analyze other challenging, overlapping subcellular structures, such as mitochondrial networks, the Golgi apparatus, or endoplasmic reticulum subdomains in various cell types. It is particularly well-suited for quantitative phenotyping in genetic or chemical screens where precise morphological measurements are required.
Materials and reagents
Biological materials
1. Saccharomyces cerevisiae strains, SK1 background [6]
a. WT strain, YSIY1218
MATa/MATα his3ΔSK/his3ΔSK ura3/ura3::pRS306-mCherry-Spo2051–91 trp1::hisG/trp1::hisG leu2/leu2 arg4-NspI/ARG4 lys2/lys2 ho∆:: LYS2/ho∆:: LYS2 rme1::LEU2/RME1 AUR1::PACT1-LexA-ER-haVP16::AUR1-C/AUR1::PACT1-LexA-ER-haVP16::AUR1-C
NDT80::hphNT1::P4×lexA-9×Myc-NDT80 /NDT80::hphNT1::P4×lexA-9×Myc-NDT80
b. gip1∆ strain, YSIY793
MATa/MATα his3ΔSK/his3ΔSK ura3/ura3::pRS306-mCherry-Spo2051–91 trp1::hisG/trp1::hisG leu2/leu2 arg4-NspI/ARG4 lys2/lys2 ho∆: :LYS2/ho∆: :LYS2 rme1::LEU2/RME1
AUR1::PACT1-LexA-ER-haVP16::AUR1-C/AUR1::PACT1-LexA-ER-haVP16::AUR1-CNDT80::hphNT1::P4×lexA-9×Myc-NDT80 /NDT80::hphNT1::P4×lexA-9×Myc-NDT80 gip1∆:: kanMX6/gip1∆:: kanMX6
2. The plasmid used in this study [6] was pRS306-mCherry-Spo2051–91URA3, integration, pTEF1-mCherry-Spo2051–91
Reagents
1. Yeast nitrogen base without amino acids (Difco YNB, catalog number: 291940)
2. D(+)-glucose (FUJIFILM Wako Pure Chemical Corporation, catalog number: 045-31167)
3. STAR agar L-grade 01 (Rikaken Co., catalog number: RSU-AL01-500G)
4. Bacto peptone (Thermo Fisher Scientific, catalog number: 211677)
5. Bacto yeast extract (Thermo Fisher Scientific, catalog number: 212750)
6. Adenine (6-Aminopurine) (FUJIFILM Wako Pure Chemical Corporation, catalog number: 012-11512)
7. Potassium acetate (FUJIFILM Wako Pure Chemical Corporation, catalog number: 167–3185)
8. β-Estradiol (Sigma-Aldrich, catalog number: E8875)
9. Amino acid mixture (five amino acid drop-out: arginine, leucine, uracil, histidine, tryptophan) (mixed manually in our lab)
Solutions
1. SD-RLU (see Recipes)
2. YPD medium (see Recipes)
3. YPA medium (see Recipes)
4. Sporulation medium (see Recipes)
5. Amino acid mixture (see Recipes)
6. 2 mM β-Estradiol (see Recipes)
Recipes
1. SD-RLU
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Yeast nitrogen base without amino acids | 6.7 g/L | 3.35 g |
| Amino acid mixture | 1 g/L | 0.5 g |
| L-Histidine | 0.16 g/L | 0.08 g |
| L-Tryptophan | 0.16 g/L | 0.08 g |
| 20% D(+)-glucose solution | 2% D(+)-glucose | 50 mL |
| Deionized water | n/a | 450 mL |
| Total | n/a | 500 mL |
a. Dissolve all ingredients except 20% D(+)-glucose solution in 450 mL of deionized water.
b. Adjust the pH to 6.5 if necessary and then autoclave.
c. Allow medium to cool to ~55 °C and then add 50 mL of 20% dextrose (glucose) solution to 2% final concentration.
d. For making plates, add STAR agar L-grade 01 (2% at final concentration) before autoclaving.
2. YPD medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Bacto yeast extract | 10 g/L | 5 g |
| Bacto peptone | 20 g/L | 10 g |
| Adenine | 0.03 g | 0.015 g |
| 20% D(+)-glucose solution | 2% D(+)-glucose | 50 mL |
| Deionized water | n/a | 450 mL |
| Total | n/a | 500 mL |
a. Dissolve all ingredients except 20% D(+)-glucose solution in 450 mL of deionized water and then autoclave.
b. Allow medium to cool to ~55 °C and add 50 mL of 20% dextrose (glucose) solution to 2% final concentration.
c. For making plates, add STAR agar L-grade 01 (2% at final concentration) before autoclaving.
3. YPA medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Bacto yeast extract | 10 g/L | 5 g |
| Bacto peptone | 20 g/L | 10 g |
| Adenine | 0.03 g | 0.015 g |
| 20% potassium acetate solution | 2% potassium acetate | 50 mL |
| Deionized water | n/a | 450 mL |
| Total | n/a | 500 mL |
a. Dissolve all ingredients except the potassium acetate solution in 450 mL of deionized water and then autoclave.
b. Allow medium to cool to ~55 °C and add 50 mL of 20% potassium acetate solution to 2% final concentration.
4. Sporulation medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 20% potassium acetate solution | 2% potassium acetate | 50 mL |
| Deionized water | n/a | 450 mL |
| Total | n/a | 500 mL |
a. Dissolve all ingredients and autoclave.
5. Amino acid mixture
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Adenine (012-11512) | n/a | 0.5 g |
| L-Alanine (010-01042) | n/a | 2 g |
| L(+)-Arginine (017-04612) | n/a | (2 g)* |
| L-asparagine monohydrate (019-04812) | n/a | 2 g |
| DL-aspartic acid (010-04842) | n/a | 2 g |
| L-Cysteine hydrochloride monohydrate (033-05272) | n/a | 2 g |
| L(+)-Glutamine (074-00522) | n/a | 2 g |
| L-Glutamic acid hydrochloride (071-02092) | n/a | 2 g |
| Glycine (073-00732) | n/a | 2 g |
| L-Histidine (084-00682) | n/a | (2 g)* |
| myo-Inositol (092-00282) | n/a | 2 g |
| L(+)-Isoleucine (121-00862) | n/a | 2 g |
| L-Leucine (124-00852) | n/a | (10 g)* |
| L(+)-Lysine monohydrochloride (121-01462) | n/a | 2 g |
| L-Methionine (133-01602) | n/a | 2 g |
| p-Aminobenzoic acid (015-2332) | n/a | 0.2 g |
| L(-)-Phenylalanine (161-01302) | n/a | 2 g |
| L(-)-Proline (161-04602) | n/a | 2 g |
| L-Serine (199-00402) | n/a | 2 g |
| L(-)-Threonine (204-01322) | n/a | 2 g |
| L-Tryptophan (204-03382) | n/a | (2 g)* |
| L-Tyrosine (202-03562) | n/a | 2 g |
| Uracil (212-00062) | n/a | (2 g)* |
| L-Valine (228-00082) | n/a | 2 g |
*Quantities enclosed in parentheses indicate dropouts.
All amino acids are from FUJIFILM Wako Pure Chemical Corporation.
6. 2 mM β-Estradiol
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| β-Estradiol | 2 mM | n/a |
| Ethanol | n/a |
a. Dissolve β-Estradiol in 100% ethanol to a final concentration of 2 mM.
b. Aliquot into 1.5 mL tubes and store at -30 °C.
Laboratory supplies
1. Sterile Veritable Petri dishes (Shallow, catalog number: 36-3412)
2. 24 × 50 mm cover slip (Matsunami Glass Ind., Ltd., No.1, catalog number: C024501)
3. 18 × 18 mm cover slip (Matsunami Glass Ind., Ltd., No.1, catalog number: C218181)
4. 250 mL Erlenmeyer flask (Corning, catalog number: 431144)
5. Incubator (PHC Holdings Corporation, catalog number: MIR-154-PJ)
6. Shaking water bath (Personal-11, TAITEC CORPORATION, catalog number: 0069409-000)
7. Clean bench (PHC Holdings Corporation, catalog number: MCV-131BNF)
8. Vortex mixer (LMS Laboratory and Medical Supplies, Brigachtal, catalog number: VTX-3000L)
9. Low speed benchtop centrifuge (TOMY, catalog number: LC-200)
Equipment
1. THUNDER Imager Live Cell system (Leica Microsystems; includes a DFC9000 GTC sCMOS camera)
2. Workstation for computation (HP, OMEN Desktop; equipped with an NVIDIA GeForce RTX 3060 Ti GPU)
3. iPad Air (5th generation) (Apple Inc., model: MM9C3J/A)
4. Apple Pencil (2nd generation) (Apple Inc., model: A2051)
Software and datasets
1. CellPose 3.1.0 (Oct 30, 2024, BSD, free)
2. MMdetection, v3.3.0 (Jan 5, 2024, Apache, free)
3. Fiji/ImageJ, 2.16.0/1.54p (Feb 17, 2025, GPL, free)
4. spacedesk Driver, 2.1.43 free (non-commercial private license)
5. Docker Engine, 27.1.4 (Dec 17, 2024, Apache, free)
Procedure
文章信息
稿件历史记录
提交日期: Aug 25, 2025
接收日期: Oct 22, 2025
在线发布日期: Nov 4, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Taguchi, S., Chagi, K., Kawai, H., Irie, K. and Suda, Y. (2025). Detailed Protocol for Segmentation and Quantification of Overlapping Prospore Membranes using DeMemSeg. Bio-protocol 15(23): e5520. DOI: 10.21769/BioProtoc.5520.
分类
生物信息学与计算生物学
细胞生物学 > 细胞结构 > 细胞质膜
微生物学 > 微生物生物膜 > 生物膜检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link