- EN - English
- CN - 中文
Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking
基于融合引物的 Racket PCR 基因组步移技术的实现
发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5517 浏览次数: 941
评审: Shengze YaoAnonymous reviewer(s)

相关实验方案

通过简并PCR鉴定二倍体马铃薯Solanum okadae中的S位点F-box蛋白序列
Amar Hundare [...] Timothy P. Robbins
2025年06月05日 2005 阅读
Abstract
Genome-walking protocols have been extensively used to clone unknown genomic sequences next to known DNAs. Existing genome-walking protocols need further improvement in methodological specificity or operation. Here, we describe a novel genome-walking protocol based on fusion primer–driven racket PCR (FPR-PCR). FPR-PCR involves four sequence-specific oligos (SSO), SSO1, SSO2, SSO3, and SSO4, which are sequentially chosen from known DNA in the direction 5’→3’. The fusion primer, mediating primary FPR-PCR, is generated by attaching SSO3 to the 5’ end of SSO1. The SSO3 encourages the target DNA of primary PCR to form a racket-like structure by mediating intra-strand annealing. SSO2 and SSO4 are directly used as sequence-specific primers (SSP) in secondary FPR-PCR, which selectively amplifies this racket-like DNA. This protocol was verified by cloning several unknown genomic sequences. Compared to traditional PCRs, FPR-PCR offers the advantages of higher specificity and fewer rounds, primarily attributed to the omission of arbitrary walking primers typically required in traditional methods.
Key features
• This FPR-PCR protocol builds upon the method constructed by Pei et al. [1].
• The FPR-PCR protocol relies on a multi-functional fusion primer (FP) that mediates the primary amplification and the formation of racket-like DNA.
• The FPR-PCR comprises only two rounds of amplification reactions.
Keywords: Genome-walking PCR (基因组步移 PCR)Graphical overview
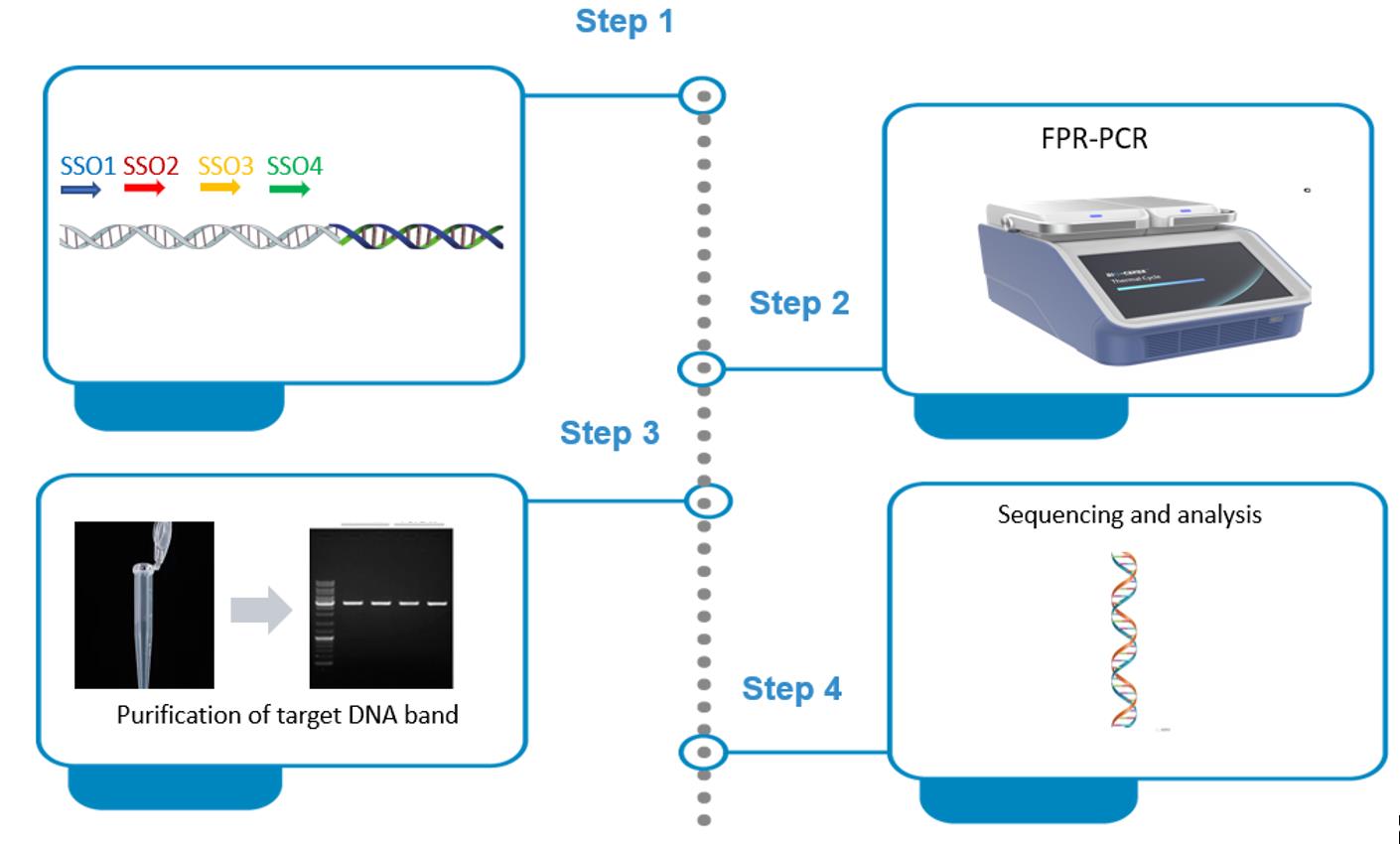
Background
Genome walking (GW) is a molecular technique for identifying unknown genomic sequences next to known DNAs [2–4]. GW has been extensively used to obtain regulatory sites of genes, amplify non-conserved regions based on conserved DNAs, identify T-DNA, discover new functional genes, or screen microbes [5–7]. Therefore, GW has made significant contributions to the development of life sciences [8–10].
To date, there are many protocols for genome walking, mainly including genomic library methods, ligation-based PCRs, and arbitrary PCRs [11–14]. The genomic library method has been abandoned due to its time-consuming nature [15]. The ligation-based PCRs are also showing a tendency to be phased out, as they require pretreatment of the genomic plate prior to PCR [16–19]. In contrast, arbitrary PCRs directly mediate walking by randomly annealing a walking primer to the unknown flank; in general, the target DNA is obtained after two to three nested amplifications [20–23]. Therefore, arbitrary PCRs are a more rapid and straightforward approach. However, existing arbitrary PCRs typically realize GW by the differential amplification between target DNA and non-target DNA. Obviously, non-target background arising from walking primer challenges these PCRs [24–26].
Herein, we propose an efficient but specific genome-walking protocol, fusion primer–driven racket PCR (FPR-PCR). This method utilizes a fusion primer (FP) to mediate the target amplicon of primary FPR-PCR to form a racket-like structure; then, a secondary PCR, driven by a sequence-specific primer (SSP) pair, selectively enriches this racket-like DNA. As a result, non-target amplification is basically overcome in FPR-PCR. The FPR-PCR was validated by successfully acquiring several unknown flanking genomic DNAs [27–29].
Materials and reagents
Biological materials
1. Genome of Levilactobacillus brevis [30–35], isolated with the TIANamp Bacteria DNA kit
Reagents
1. TIANamp Bacteria DNA kit (TIANGEN, catalog number: 4992448)
2. 1× TE buffer (Sangon, catalog number: B548106)
3. LA Taq polymerase (Takara, catalog number: RR02MA)
4. 6× Loading buffer (Takara, catalog number: 9156)
5. DiaSpin column DNA Gel Extraction kit (Sangon, catalog number: B110092)
6. DL 5,000 DNA marker (Takara, catalog number: 3428Q)
7. Agarose (Sangon, catalog number: A620014)
8. 0.5 M EDTA (Solarbio, catalog number: B540625)
9. GoldView I nucleic acid staining agent (10,000 ×) (Solarbio, catalog number: G8140)
10. Tris (Solarbio, catalog number: T8060)
11. Boric acid (Solarbio, catalog number: B8110)
12. Oligos (Sangon)
gadC-FPα: TGTTTTCTTCTTGCTCT|ATGGTTATTCTCTGGGG
gadC-FPβ: TGTTTTCTTCTTGCTCT|TCTCTGGGGATTGATTG
gadC-SSP2: TTGGGCGTTATAATTCCTGTTTTCTTCTTG
gadC-SSP4: GGAGCGGTAGTGTGTTAGTTGGGTT
Note: The two parts (SSO1 and SSO3) in an FP are separated by a vertical line. The left part is SSO3, and the right part is SSO1.
Solutions
1. 100 μM primer (see Recipes)
2. 10 μM primer (see Recipes)
3. 2.5× TBE buffer (see Recipes)
4. 0.5× TBE buffer (see Recipes)
5. 1.5% agarose gel (see Recipes)
Recipes
1. 100 μM primer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Primer powder | 100 μM | n/a |
| 1× TE buffer | 1× | Volume (μL) specified by the supplier |
| Total | n/a | Volume (μL) specified by the supplier |
2. 10 μM primer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 100 μM primer | 10 μM | 10 μL |
| Ultrapure water | n/a | 90 μL |
| Total | n/a | 100 μL |
3. 2.5× TBE buffer (pH 8.3)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Tris | 225 mM | 27 g |
| Boric acid | 225 mM | 13.75 g |
| 0.5 M EDTA | 5 mM | 10 mL |
| Ultrapure water | n/a | 950 mL |
| Total | n/a | 1,000 mL |
4. 0.5× TBE buffer (pH 8.3)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 2.5 × TBE buffer | 0.5× | 200 mL |
| Ultrapure water | n/a | 800 mL |
| Total | n/a | 1,000 mL |
5. 1.5% agarose gel
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Agarose powder | 1.5% | 1.5 g |
| 0.5× TBE buffer | 0.5× | 100 mL |
| GoldView I nucleic acid staining agent (10,000×) | 1× | 10 μL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 0.2 mL PCR tubes (Kirgen, catalog number: KG2311)
2. 10 μL pipette tips (Sangon, catalog number: F600215)
3. 0.2 mL pipette tips (Sangon, catalog number: F600227)
4. 1 mL pipette tips (Sangon, catalog number: F630101)
5. 1.5 mL tubes (Labselect, catalog number: MCT-001-150)
Equipment
1. PCR cycler (Analtytikjena, model: Biometra TOne 96G PCR)
2. Electrophoresis apparatus (Beijing Liuyi, model: DYY-6C)
3. Gel imaging system (Bio-Rad, model: ChemiDoc XRS+)
4. Microcentrifuge (Tiangen, model: TGear)
Software and datasets
1. Oligo 7 software (Molecular Biology Insights, Inc., USA)
2. DNASTAR Lasergene software (DNASTAR, Inc., USA)
3. All data are available at https://www.frontiersin.org/articles/10.3389/fgene.2022.969840/full#supplementary-material (access date, 10/18/2022)
Procedure
文章信息
稿件历史记录
提交日期: Sep 22, 2025
接收日期: Oct 24, 2025
在线发布日期: Nov 3, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Gu, Y., Pei, J., Li, M., Tang, Q. and Li, H. (2025). Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking. Bio-protocol 15(23): e5517. DOI: 10.21769/BioProtoc.5517.
分类
分子生物学 > DNA > 基因组步移
微生物学 > 微生物遗传学 > DNA
分子生物学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










