- EN - English
- CN - 中文
Analyzing the Translatome of Lymphatic and Venous Endothelial Cells In Vivo via Translating Ribosome Affinity Purification (TRAP)
利用翻译核糖体亲和纯化(TRAP)技术解析体内淋巴与静脉内皮细胞的转录后表达谱
(*contributed equally to this work) 发布: 2025年12月05日第15卷第23期 DOI: 10.21769/BioProtoc.5516 浏览次数: 1672
评审: Alberto RissoneAmr Galal Abdelraheem IbrahimAnonymous reviewer(s)
Abstract
Zebrafish are a powerful model for investigating vascular and lymphatic biology due to their genetic tractability and optical transparency. While translating ribosome affinity purification (TRAP) has been widely applied in other systems, its application in zebrafish has remained limited. Here, we present an optimized TRAP protocol for isolating ribosome-associated mRNAs from endothelial cells in vivo, without the need for cell dissociation or sorting. Using a novel transgenic zebrafish line, which expresses HA-tagged Rpl10a under the mrc1a promoter, we enriched actively translating endothelial transcripts. Differential expression analysis revealed robust upregulation of vascular and lymphatic genes including flt4, kdrl, and lyve1b. This approach captures the endothelial cell translatome with high specificity and offers a robust platform for investigating the molecular mechanisms of endothelial biology under genetic, environmental, or toxicological perturbations.
Key features
• Permits in vivo isolation of the endothelial ribosome without cell sorting.
• Optimized TRAP protocol to analyze lymphatic and venous endothelial gene expression in zebrafish.
• Utilizes a stable transgenic zebrafish line.
• Compatible with real-time quantitative PCR (qPCR) and next-generation sequencing (RNA-seq).
Keywords: TRAP (TRAP)Graphical overview
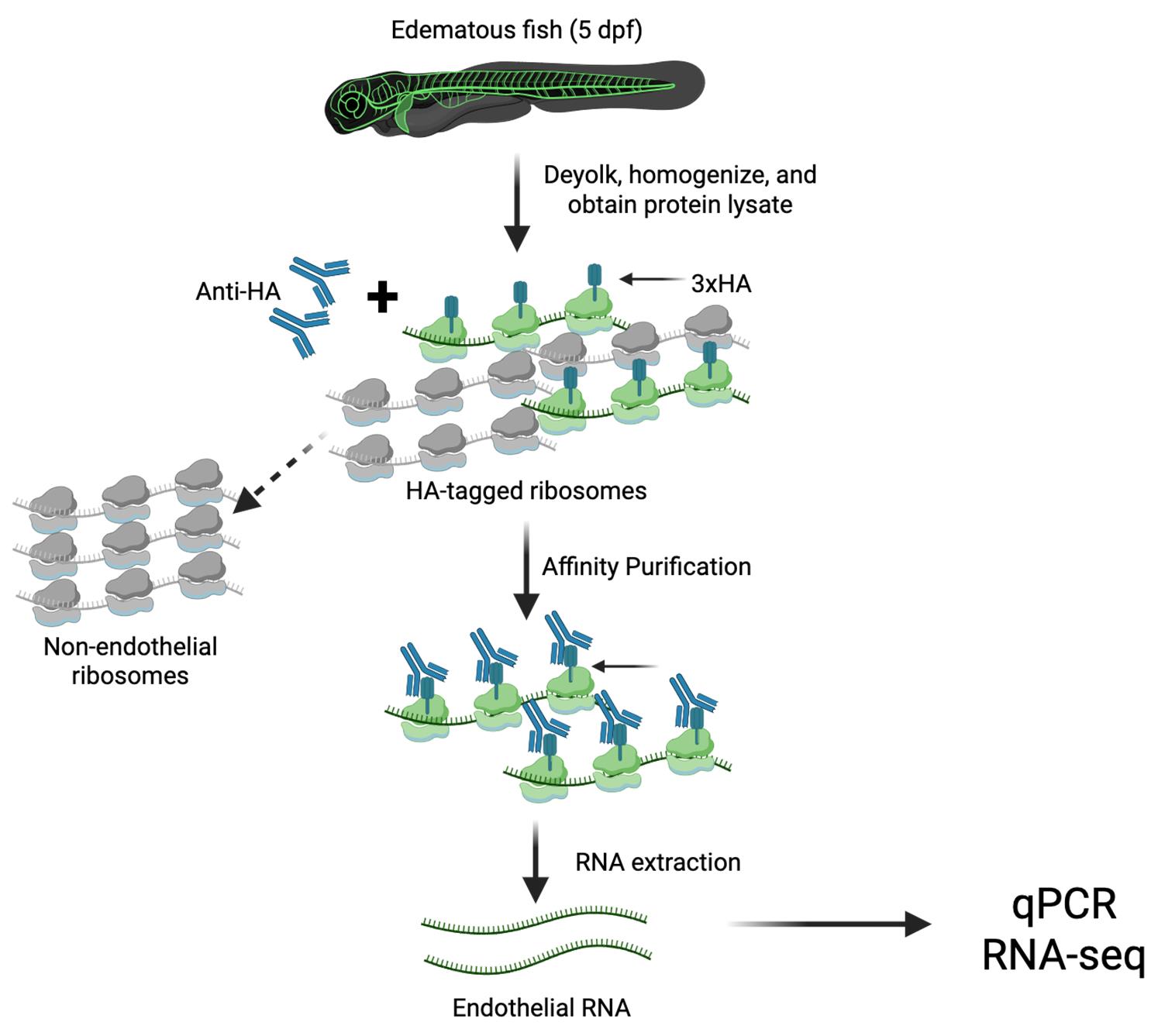
Graphical overview of the translating ribosome affinity purification (TRAP) protocol. Transgenic RiboTag zebrafish are deyolked and homogenized to generate whole-tissue lysates. Anti-HA antibodies are added to the lysate to selectively bind HA-tagged endothelial ribosomes. Magnetic Dynabeads conjugated to the antibody Fc domain enable affinity purification of the HA-tagged ribosomes using a magnetic stand. Unbound, non-endothelial ribosomes remain in the supernatant, while the Dynabead–αHA-ribosome–mRNA complex is subjected to RNA extraction to isolate actively translated endothelial RNA. This protocol provides an in vivo snapshot of cell type–specific translational activity.
Background
The blood and lymphatic systems form an integrated vascular network that delivers oxygen and nutrients, removes metabolic waste, and maintains fluid balance and homeostasis. These networks are composed of endothelial cells, whose functions are tightly regulated under both physiological and pathological conditions [1,2]. Disruption of endothelial regulation can impair fluid balance, resulting in fluid accumulation and tissue edema. In response to edemagenic signals in the microenvironment, lymphatic and venous endothelial cells activate molecular programs that promote edema clearance [3].
Zebrafish have emerged as a valuable animal model for studying vascular development due to their optical transparency, genetic tractability, and evolutionary conservation of endothelial gene function [4–7]. In our recent study, we utilized live imaging of early-stage zebrafish larvae exposed to abrupt salinity changes to induce osmotic stress and tissue edema. This model revealed increased lymphangiogenesis and remodeling of the lymphatic vasculature during edema resolution, characterized by proliferation of lymphatic progenitors and expansion of the primary lymphatic network [3].
Although endothelial responses to edema are generally initiated at the transcriptional level, post-transcriptional mechanisms, including translational regulation, likely contribute to dynamic changes in gene expression [8,9]. Transcriptome profiling tools such as microarrays and RNA sequencing from total RNA provide insights into RNA abundance, but they do not reflect the state of translation activity [10,11]. Since protein synthesis is determined by mRNAs engaged with actively translating ribosomes, profiling the translatome offers a more accurate representation of functional gene expression.
Conventional transcriptome analyses rely on fluorescence-activated cell sorting (FACS) of labeled endothelial cells, which requires mechanical and chemical dissociation into single-cell suspensions [12,13]. However, this process can disrupt native cell–cell and cell–matrix interactions, potentially altering gene expression and introducing artifacts. Translating ribosome affinity purification (TRAP) using the RiboTag strategy provides a powerful alternative for cell type–specific isolation of ribosome-associated mRNAs in vivo, bypassing the need for cell sorting [14–20]. The rapid, non-disruptive nature of TRAP minimizes ex vivo artifacts and preserves in vivo gene expression profiles.
To enable endothelial-specific TRAP in zebrafish, we developed a transgenic line Tg(mrc1a:egfp-2a-rpl10a-3xHA)y723, expressing hemagglutinin (HA)-tagged Rpl10a and eGFP via a viral 2A peptide under control of the endothelial-specific mrc1a promoter [3]. Following osmotic challenge, larvae display vascular remodeling and enable the isolation of actively translating mRNAs from lymphatic and venous endothelial cells. Translatome analysis revealed upregulation of key endothelial genes such as flt4, kdrl, and lyve1b [3]. Coupling TRAP with RNA sequencing allows for comprehensive identification of genes involved in endothelial responses to edema.
This protocol describes an optimized TRAP workflow for profiling the endothelial translatome in edematous zebrafish. It provides a robust, reproducible method for capturing dynamic gene expression changes during edema formation and resolution.
Materials and reagents
Biological materials
1. Stable transgenic zebrafish line: Tg(mrc1a:egfp-2a-rpl10a-3xHA)y723
Reagents
1. Invitrogen DynabeadsTM Protein G (ThermoFisher, catalog number: 10004D)
2. Direct-zolTM RNA MicroPrep (ZymoResearch, catalog number: R2060)
3. 100 μg/mL cycloheximide (Sigma, catalog number: C7698)
4. Protease inhibitor cocktail (Sigma, catalog number: P8340)
5. PierceTM Bradford Protein Assay kit (ThermoFisher, catalog number: 23200)
6. 1 mM DTT (Sigma, catalog number: 646563)
7. 200 units/mL RNAsin (Promega, catalog number: N2115)
8. 1 mg/mL heparin (Sigma, catalog number: H3393-10KU)
9. 10% NP-40 (Roche, catalog number: 11-332-473-001)
10. Anti-HA antibody (Abcam, catalog number: ab9110)
11. TRIzol (Ambion Life Technologies, catalog number: 15596026)
12. Glycogen RNA-grade (ThermoFisher, catalog number: R0551)
13. High-Capacity cDNA Reverse Transcription kit (Applied Biosystem, catalog number: 4374966)
14. TaqMan Fast Advanced Master Mix (Applied Biosystem, catalog number: 4444557)
15. Phosphate-buffered saline (PBS), pH 7.4 (Fisher, catalog number: BP2438)
16. Pronase from Streptomyces griseus (20 mg/mL) (Sigma, catalog number: 10165921001)
17. Ethyl alcohol 200 proof (Sigma, catalog number: E7023-1L)
18. Chloroform HPLC-grade (Fisher, catalog number: C606SK-1)
19. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014-1KG)
20. Potassium chloride (KCl) (Fisher Chemical, catalog number: P217-500)
21. Sodium bicarbonate (NaHCO3) (Fisher Chemical, catalog number: S233-500)
22. Calcium chloride (CaCl2) (MP Biomedicals, catalog number: 193819)
23. Magnesium chloride (MgCl2) (Invitrogen, catalog number: AM9530G)
24. Calcium nitrate hydrate [Ca(NO3)2·xH2O] (Alfa Aesar, catalog number: 44515-14)
25. Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391-25G)
26. Tris pH 8.5 (Teknova, catalog number: 21327)
27. Tris-HCl pH 7.4 (Teknova, catalog number: T1074)
28. Molecular grade water (H2O) (Corning, catalog number: 46-000-CM)
29. HEPES (Fisher Bioreagents, catalog number: BP310-500)
Solutions
1. Deyolking buffer (see Recipes)
2. Deyolking wash buffer (see Recipes)
3. Homogenization buffer (HB) (see Recipes)
4. Homogenization buffer+ (HB+) (see Recipes)
5. High salt buffer (HSB) (see Recipes)
6. High salt buffer+ (HSB+) (see Recipes)
7. Hypertonic salt solution (3× Danieau buffer) obtained from 10× Danieau buffer stock (see Recipes)
8. High-capacity RT mix for cDNA synthesis (see Recipes)
9. TaqMan Real-Time PCR (see Recipes)
Recipes
1. Deyolking buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M NaCl | 55 mM | 5.50 mL |
| 1 M KCl | 1.9 mM | 0.19 mL |
| 1 M NaHCO3 | 1.25 mM | 0.125 mL |
| H2O | n/a | 94.185 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
2. Deyolking wash buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M NaCl | 110 mM | 11.00 mL |
| 1 M KCl | 3.5 mM | 0.35 mL |
| 1 M CaCl2 | 2.7 mM | 0.27 mL |
| 1 M Tris (pH 8.5) | 10 mM | 1.00 mL |
| H2O | n/a | 87.38 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
3. Homogenization buffer (HB)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris-HCl (pH 7.4) | 50 mM | 5.00 mL |
| 1 M KCl | 100 mM | 10.00 mL |
| 1 M MgCl2 | 12 mM | 1.20 mL |
| 10% NP-40 | 1% | 10 mL |
| H2O | n/a | 73.80 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
4. Homogenization buffer+ (HB+)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DTT | 1 mM | 10 μL |
| Protease inhibitor cocktail | 0.01× | 100 μL |
| RNAsin | 200 units/mL | 50 μL |
| Cycloheximide | 100 μg/mL | 50 μL |
| Heparin | 1 mg/mL | 100 μL |
| HB (see Recipe 3) | n/a | 9.69 mL |
| Total | n/a | 10 mL |
Freshly prepared on ice.
5. High salt buffer (HSB)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Tris (pH 7.4) | 50 mM | 5.00 mL |
| 1 M KCl | 300 mM | 30.00 mL |
| 1 M MgCl2 | 12 mM | 1.20 mL |
| 10% NP-40 | 1% | 10.00 mL |
| H2O | n/a | 53.80 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
6. High salt buffer+ (HSB+)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DTT | 1 mM | 10 μL |
| Protease inhibitor cocktail | 0.01× | 100 μL |
| RNAsin | 200 units/mL | 50 μL |
| Cycloheximide | 100 μg/mL | 50 μL |
| Heparin | 1 mg/mL | 100 μL |
| HSB (see Recipe 5) | n/a | 9.69 mL |
| Total | n/a | 10 mL |
Freshly prepared on ice.
7. Hypertonic salt solution (3× Danieau buffer) obtained from 10× Danieau buffer stock
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 580 mM NaCl | 58 mM | 10.00 mL |
| 6.71 mM KCl | 0.7 mM | 10.43 mL |
| 8.22 mM MgSO4 | 0.4 mM | 4.87 mL |
| 8.53 mM Ca(NO3)2 | 0.6 mM | 7.03 mL |
| 50 mM HEPES | 10 mM | 20.0 mL |
| H2O | n/a | 47.67 mL |
| Total | n/a | 100 mL |
Store at 4 °C.
8. High-capacity RT mix for cDNA synthesis
| Component | Quantity or volume |
|---|---|
| 10× RT buffer | 2.0 µL |
| 25× dNTP Mix | 0.8 µL |
| 10× RT random primers | 2.0 µL |
| MultiScribeTM reverse transcriptase | 1.0 µL |
| RNase inhibitor | 1.0 µL |
| Nuclease-free H2O | 3.2 µL |
| RNA template (100 ng) | 10 µL |
| Total per reaction | 20 µL |
Volumes per reaction. Store at -20 °C.
9. TaqMan real-time PCR
| Component | Quantity or volume |
|---|---|
| TaqMan Fast Advanced Master Mix (2×) | 5.0 µL |
| TaqMan target gene primers FAM (20×) | 0.5 µL |
| TaqMan reference gene primers VIC (20×) | 0.5 µL |
| Nuclease-free H2O | 2.0 µL |
| cDNA (1:5) | 2.0 µL |
| Total per reaction | 10 µL |
Volumes per reaction. Store at -20 °C.
Laboratory supplies
1. 1.5 mL microcentrifuge tubes (ABDOS, catalog number: P10202)
2. RNase-free elution tubes (Invitrogen, catalog number: AM12480)
3. VWR disposable pestles (VWR, catalog number: 47747-358)
4. SureOne sterile pipette tips (FisherBrand, catalog number: 02-707-442)
Equipment
1. Magnetic Eppendorf stand (ThermoFisher, catalog number: 12321D)
2. SorvallTM LegendTM Micro 21R Microcentrifuge 4 °C microfuge (ThermoFisher, catalog number: 75002445)
3. 4 °C Gentle Eppendorf tube rotator (Fisher, catalog number: 11-676-342)
4. Zymo-DR Duet kit (ZymoResearch, catalog number: R2060)
5. Labnet centrifuge (Thomas Scientific, catalog number: 1160W29)
6. Vortex (Fisher, catalog number: 88-882-011)
7. Zeiss Stemi 305 microscope (Zeiss, catalog number: 435063-9020-100)
8. Biovortexer (BioSpec, model: 1083MC)
9. NanoDrop (Thermo Scientific, catalog number: 13400518)
10. Confocal laser scanning microscope (Zeiss, model: LSM880)
11. Quantitative PCR machine (Applied Biosystems, model: Quant Studio 7 Flex)
Software and datasets
1. GraphPad Prism Software, version 9.2.0
Procedure
文章信息
稿件历史记录
提交日期: Aug 22, 2025
接收日期: Oct 22, 2025
在线发布日期: Nov 3, 2025
出版日期: Dec 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Olayinka, O., Zarinebaf, L. and Jung, H. M. (2025). Analyzing the Translatome of Lymphatic and Venous Endothelial Cells In Vivo via Translating Ribosome Affinity Purification (TRAP). Bio-protocol 15(23): e5516. DOI: 10.21769/BioProtoc.5516.
分类
分子生物学 > RNA > mRNA 转译
细胞生物学 > 细胞器分离 > 多聚核糖体
发育生物学 > 细胞生长和命运决定 > 淋巴管生成
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link