- EN - English
- CN - 中文
Click-qPCR: A Simple Tool for Interactive qPCR Data Analysis
Click-qPCR:一种用于交互式 qPCR 数据分析的简便工具
发布: 2025年11月20日第15卷第22期 DOI: 10.21769/BioProtoc.5513 浏览次数: 2718
评审: Sébastien GillotinAnonymous reviewer(s)
Abstract
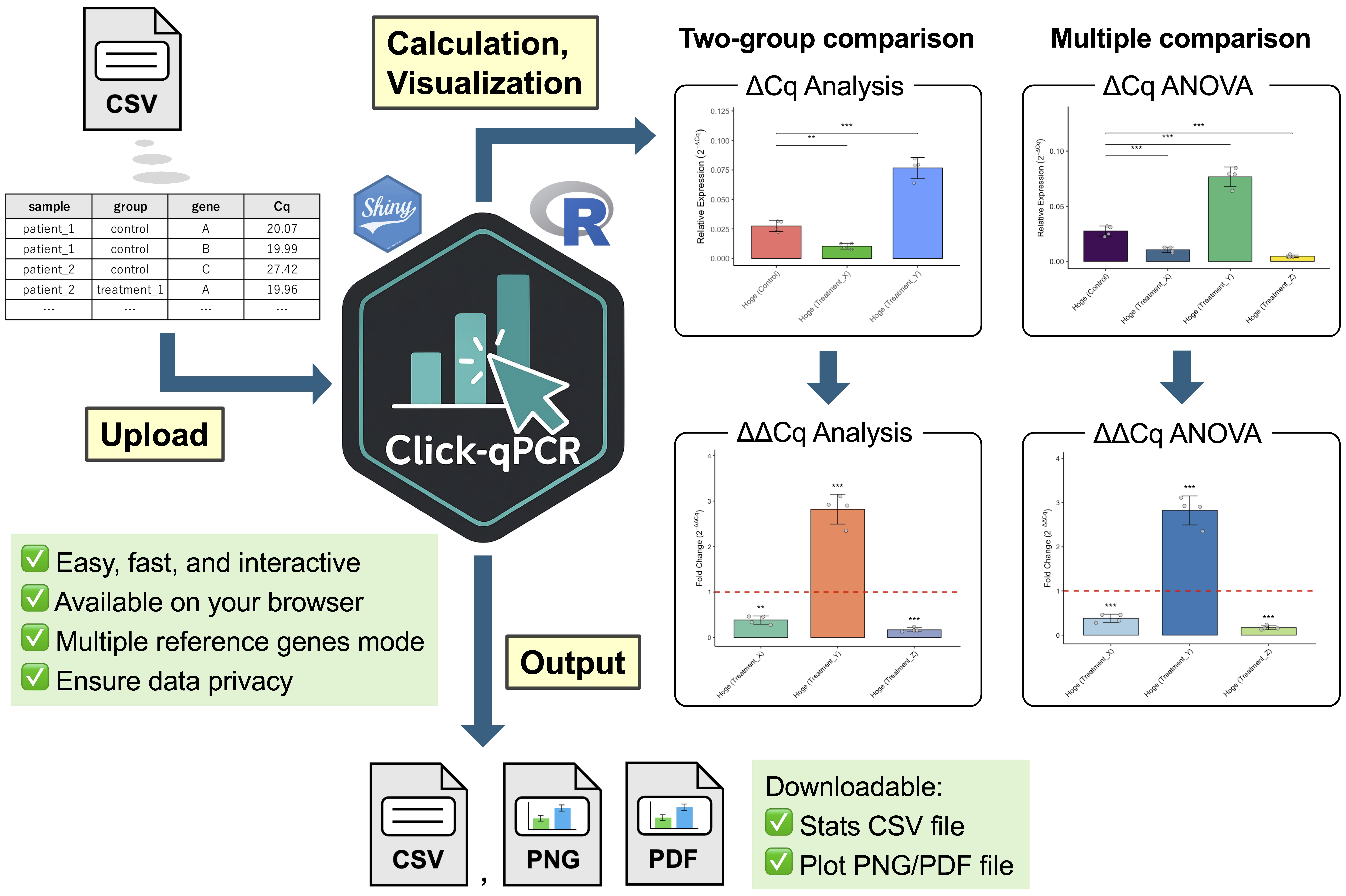
Real-time quantitative PCR (qPCR) is a pivotal technique for analyzing gene expression and DNA copy number variations. However, the limited availability of user-friendly software tools for qPCR data analysis presents a significant challenge for experimental biologists with limited computational skills. To address this issue, we developed Click-qPCR, a user-friendly and web-based Shiny application for qPCR data analysis. Click-qPCR streamlines ΔCq and ΔΔCq calculations using user-uploaded CSV data files. The interactive interface of the application allows users to select genes and experimental groups and perform Welch’s t tests and one-way analysis of variance with Dunnett’s post-hoc test for pairwise and multi-group comparisons, respectively. Results are visualized via interactive bar plots (mean ± standard deviation with individual data points) and can be downloaded as publication-quality images, along with summary statistics. Click-qPCR empowers researchers to efficiently process, interpret, and visualize qPCR data regardless of their programming experience, thereby facilitating routine analysis tasks. Click-qPCR Shiny application is available at https://kubo-azu.shinyapps.io/Click-qPCR/, while its source code and user guide are available at https://github.com/kubo-azu/Click-qPCR.
Key features
• A simple browser-based interface that enables biologists without coding experience to perform qPCR data analysis.
• Provides integrated data analysis workflows for ΔCq/ΔΔCq calculations and one-way analysis of variance, with appropriate statistical tests and generation of publication-ready visualizations.
• Designed for experimental contexts that require quick and reproducible comparisons of gene expression across various conditions, treatments, and reference genes.
• Open-source and freely accessible online, supporting transparency, reproducibility, and broad adoption within molecular biology and biomedical research communities.
Keywords: Real-time quantitative PCR (qPCR) (实时定量 PCR(qPCR))Graphical overview
Background
Real-time quantitative PCR (qPCR) is a cornerstone technique in molecular biology that enables highly sensitive profiling of gene expression levels [1]. Application areas of qPCR span diverse fields of research, ranging from basic biological investigations to clinical diagnostics. In addition to assessing mRNA expression profiles, qPCR is a robust method for detecting DNA copy number variations (CNVs). While both absolute and relative quantification strategies can be used, relative quantification is more commonly used in qPCR analysis due to the inherent complexities and specific requirements of absolute quantification, which often limit its utility in specialized applications such as virus quantification. The ΔΔCq (ΔΔCt) method commonly used for relative quantification in qPCR analysis robustly compares mRNA transcript levels and CNVs [2]. This method determines relative gene expression by first normalizing the target gene’s Cq value with respect to those of reference genes, then rescaling the normalized value to a control group. This procedure is crucial for accurately interpreting biological changes in gene expression and for CNV analysis.
Despite its widespread adoption and critical importance for relative gene expression and CNV analysis, the ΔΔCq method often presents significant hurdles for wet-lab researchers in terms of accessing and utilizing appropriate analysis software. Many available software solutions are proprietary, commercially licensed, platform-specific, or require a certain level of computational skills and programming knowledge, which can be a barrier for many experimental biologists. For these researchers, alternative solutions such as spreadsheet-based qPCR analysis are often error-prone and time-consuming for multiple genes, whereas advanced statistical software can be unnecessarily complex for routine tasks.
To overcome these limitations, we developed a user-friendly tool for qPCR data analysis: Click-qPCR. Unlike many existing solutions that may require local software installation, command-line operations, or a foundational understanding of programming environments, Click-qPCR is a web-based application that requires no setup beyond a standard internet browser. This tool is designed to provide a user-friendly, intuitive, and readily accessible web-based platform specifically designed for rapid and interactive analysis of qPCR data. The application can instantly process typical qPCR datasets, such as those from a 384-well plate, making it ideal for routine high-throughput analyses. Its focus on operational simplicity and immediate visual feedback makes Click-qPCR ideal for researchers, students, and technicians with limited bioinformatics experience.
Software and datasets
The Click-qPCR application was developed using the R programming language [3] (version 4.4.2) and Shiny framework [4] to provide an interactive web interface. It depends on several R packages publicly available from the CRAN repository. The application is platform-independent and can be run on any operating system with a standard R installation. Alternatively, users are required to install the following packages (Table 1) that can be obtained using install.packages():
```Rinstall.packages(c("shiny", "shinyjs", "readr", "dplyr", "ggplot2", "tidyr", "DT", "RColorBrewer", "fontawesome", "multcomp"))```
Table 1. Required packages for Click-qPCR application
| Type | Software/dataset/resource | Version | Date | License | Access (free or paid) |
|---|---|---|---|---|---|
| Software | R [3] | 4.4.2 | 2024.10.31 | GNU GPL | Free |
| Software | Shiny [4] | 1.10.0 | 2024.12.16 | GPL-3 | Free |
| Software | Shinyjs [5] | 2.1.0 | 2021.12.20 | MIT | Free |
| Software | Readr [6] | 2.1.5 | 2025.07.23 | MIT | Free |
| Software | Dplyr [7] | 1.1.4 | 2023.11.17 | MIT | Free |
| Software | Tidyr [8] | 1.3.1 | 2024.01.24 | MIT | Free |
| Software | ggplot2 [9] | 3.5.2 | 2025.04.09 | MIT | Free |
| Software | DT [10] | 0.33 | 2024.04.04 | MIT | Free |
| Software | RColorBrewer [11] | 1.1-3 | 2022.04.03 | Apache License 2.0 | Free |
| Software | Fontawesome [12] | 0.5.3 | 2024.11.16 | MIT | Free |
| Software | Multcomp [13] | 1.4-28 | 2025.01.29 | GPL-2 | Free |
The user guide and detailed installation instructions for the local user environment are available at https://github.com/kubo-azu/Click-qPCR. The template CSV file for Click-qPCR is available at https://github.com/kubo-azu/Click-qPCR/blob/main/Click-qPCR_template.csv.
Procedure
文章信息
稿件历史记录
提交日期: Sep 8, 2025
接收日期: Oct 14, 2025
在线发布日期: Oct 30, 2025
出版日期: Nov 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Kubota, A. and Tajima, A. (2025). Click-qPCR: A Simple Tool for Interactive qPCR Data Analysis. Bio-protocol 15(22): e5513. DOI: 10.21769/BioProtoc.5513.
分类
生物信息学与计算生物学
分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













