- EN - English
- CN - 中文
Monitoring Endocytosis of Integral Membrane Proteins Using Western Blot-Based Detection of Biotinylated Antibody Uptake
利用基于 Western blot 的生物素化抗体内吞检测法监测膜整合蛋白的内吞过程
发布: 2025年11月20日第15卷第22期 DOI: 10.21769/BioProtoc.5511 浏览次数: 2195
评审: Anonymous reviewer(s)

相关实验方案

毛细管纳米免疫实验定量分析经CD138筛选的骨髓瘤细胞蛋白
Irena Misiewicz-Krzeminska [...] Norma C. Gutiérrez
2019年06月20日 6855 阅读
Abstract
The antibody-uptake assay is a commonly used technique to monitor endocytosis of integral membrane proteins including transmembrane and glycosylphosphatidylinositol-anchored proteins (GPI-APs). The antibody-uptake assay typically involves incubating live cells with fluorophore-conjugated antibodies directed against the extracellular domain of the integral membrane protein of interest. Antibody uptake is then detected by flow cytometry or confocal microscopy. However, these detection modalities may be inaccessible to some labs or require extensive training to operate. Thus, we developed an easy and novel sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot-based approach to the antibody-uptake assay that exploits the strong affinity between biotin and streptavidin. Instead of incubating cells with fluorophore-conjugated antibodies to monitor antibody uptake, our assay involves incubating cells with biotinylated antibodies, processing the cell lysates for western blot, and probing the membrane with detectably conjugated streptavidin. From preparation to quantification, this protocol requires less hands-on time than other approaches and is amenable to small-scale drug or siRNA screens. Here, we demonstrate the utility of our approach using the well-characterized misfolded GPI-AP, YFP-tagged C179A mutant of prion protein (YFP-PrP*), as our model substrate. YFP-PrP* constitutively traffics to the plasma membrane (PM), where it binds to anti-GFP antibody, and immediately undergoes endocytosis to lysosomes. To validate our protocol, we present measurements of antibody uptake under conditions known to enhance or inhibit YFP-PrP*’s traffic to the PM. Using this assay, we present new evidence that, under certain conditions, YFP-PrP* is able to undergo degradation via a pathway that does not involve exposure on the cell surface.
Key features
• Incubate live cells with biotinylated primary antibody and a lysosomal degradation inhibitor, process lysates for western blot, and probe the blot with detectably conjugated streptavidin.
• Fast, easy, and semi-quantitative assay to test whether integral membrane proteins are degraded through pathways involving exposure on the plasma membrane.
• Conduct screens for small molecules, siRNAs, or conditions that promote or inhibit traffic of your protein of interest through the plasma membrane.
• Pair this protocol with a synchronized trafficking assay to detect changes in the rate of proteins traversing the plasma membrane.
Keywords: Antibody uptake (抗体内吞)Graphical overview
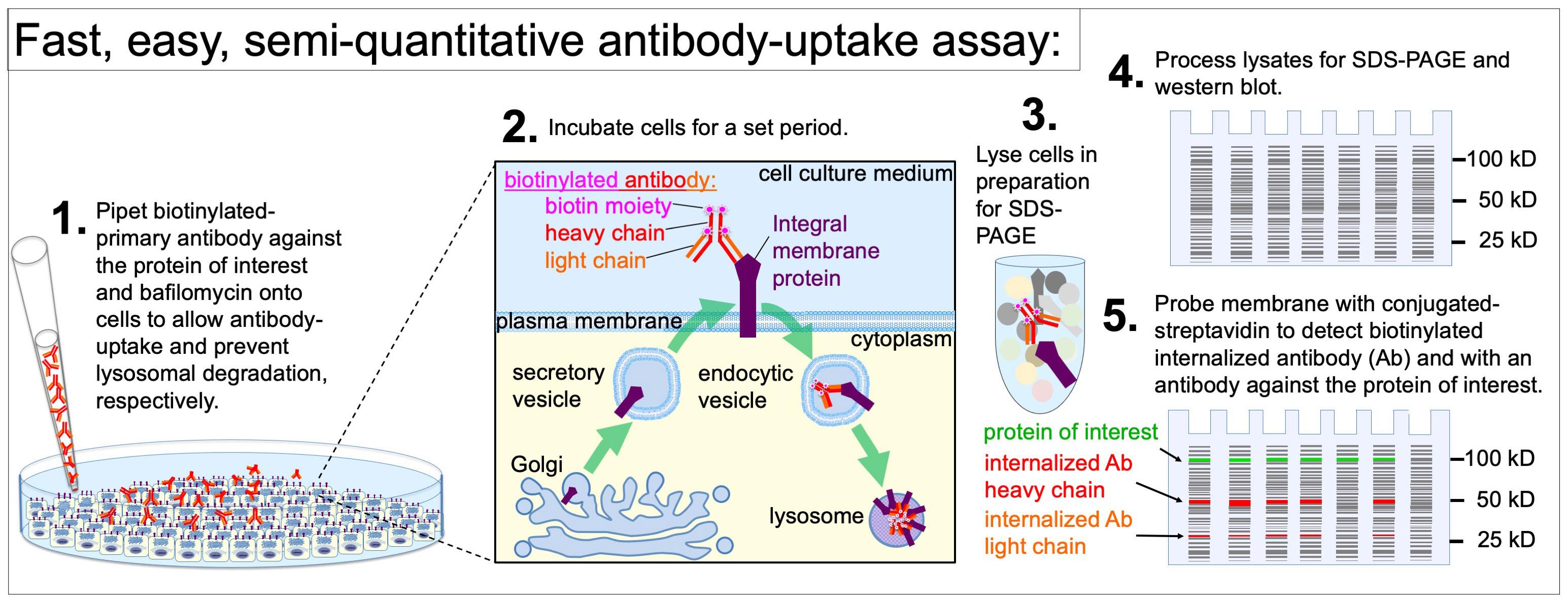
Background
History and concept of the antibody-uptake assay
Antibody uptake utilizes the specific binding of primary antibodies to membrane proteins that are displayed on the plasma membrane (PM) and subsequently internalized via endocytosis. Integral membrane proteins, including transmembrane proteins and glycosylphosphatidylinositol (GPI)-anchored proteins, are synthesized in the endoplasmic reticulum (ER) and are able to traffic from the “cradle” to the “grave” by a variety of membrane trafficking pathways (Figure 1, thin black arrows and thick dark purple arrows). Antibody-uptake assays exclusively reveal whether a population of the integral membrane proteins traffic to, and is endocytosed from, the PM (Figure 1, thick purple arrows). Determining whether a population of proteins traffic to the cell surface is essential for delineating the possible trafficking pathways an integral membrane protein may take prior to degradation. Additionally, antibody-uptake assays may inform our understanding of the signal transduction pathways that regulate the secretion and internalization of integral membrane protein receptors and channels.
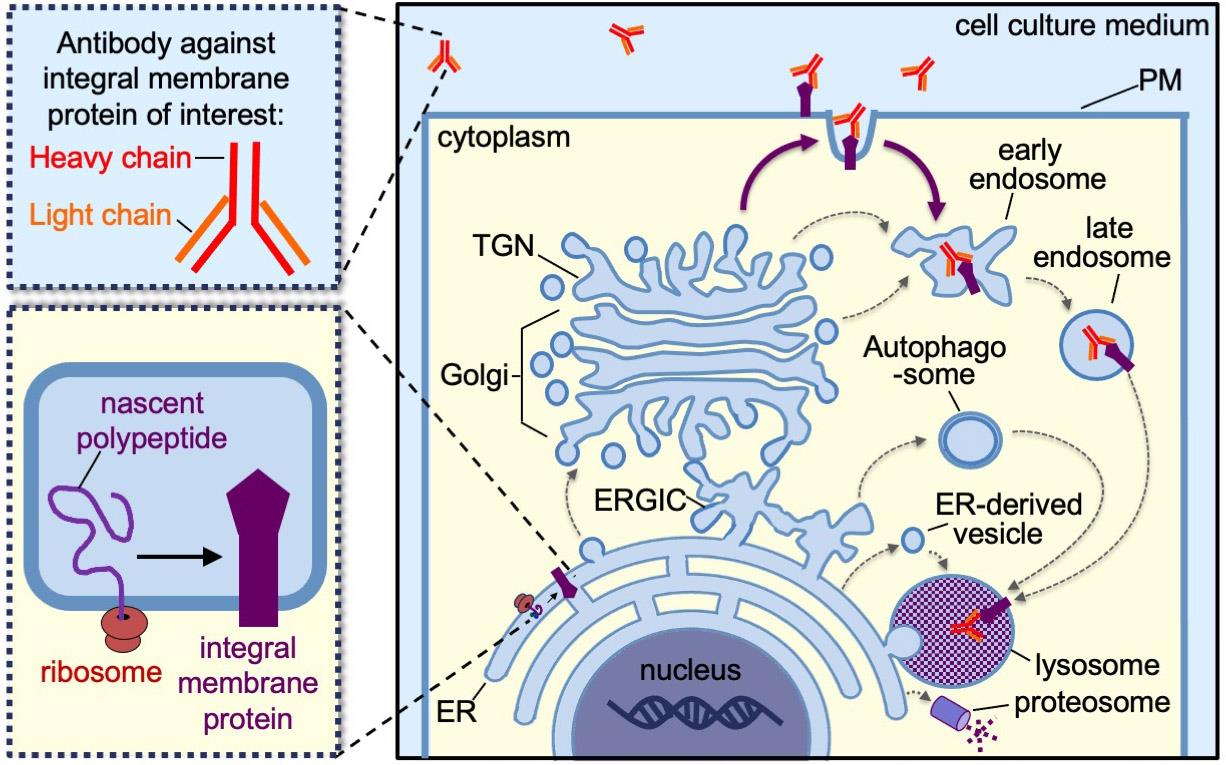
Figure 1. Schematic depicting the various trafficking pathways of an integral membrane protein from the endoplasmic reticulum, where it is synthesized (i.e., the cradle) to the lysosomes or proteosomes, where it is degraded (i.e., the grave). The thin black arrows indicate possible internal trafficking steps from one compartment to the next. The thick dark purple arrows indicate the possible trafficking steps involving transit through the PM, which specifically can be detected by antibody-uptake assays. ER, endoplasmic reticulum; ERGIC, ER–Golgi intermediate compartment; TGN, trans-Golgi network; PM, plasma membrane.
Over the past 40 years, a variety of antibody-uptake approaches have been employed to monitor the internalization of integral membrane proteins from the PM. The primary antibodies are either directly conjugated to a detectable tag (e.g., fluorophore, gold particle, radiolabel) or are unlabeled and detected by incubating lysed or permeabilized fixed cells with detectable secondary antibodies. Immunofluorescence, immunogold electron microscopy, live-cell fluorescence imaging, flow cytometry, autoradiography, and western blot have each been performed to detect the internalized primary antibodies [1–8]. Each of these approaches to detect and quantify antibody-uptake results poses distinct challenges. Quantification of fluorophore-conjugated antibody uptake using microscopy-based techniques is time-consuming. Measuring fluorescence intensity in individual cells requires imaging software for volumetric analysis and can be cost-prohibitive if a microscope is not readily available. Quantification of antibody uptake of fluorophore-conjugated antibodies by flow cytometry allows for rapid readout of large populations of cells under various conditions; however, many labs do not have ready access to a flow cytometer. Both fluorescence imaging–based and flow cytometry–based quantification may be thwarted by fluorescence quenching from fluorophore crowding or the low pH of the endocytic vesicles and lysosomes. Quantification of radioactively labeled antibody uptake by autoradiography requires designated space and equipment for radioactive work. Microscopy, flow cytometry, and radioactive work all require extensive training. By contrast, SDS-PAGE followed by western blot–based protocols using secondary antibodies to detect internalized primary antibodies utilize relatively inexpensive and standard lab equipment, require less training, and can be completed in 1 or 2 workdays. However, the standard western blot approach using primary and secondary antibodies has not provided a clear, reliable, or measurable readout for antibody uptake beyond a binary yes/no [1,7], and has not gained traction in the cell biology field. This may be in part because detection with secondary antibodies on western blot membranes depends on the internalized primary antibody maintaining antigenicity during denaturation in the lysosomes and during the subsequent cell lysis and SDS-PAGE process.
A new approach to the antibody-uptake assay
To address the practical, technical, and biological limitations of the conventional approaches to detect antibody uptake, we developed a novel approach that builds on the advantages of SDS-PAGE and western blot. Our approach exploits the extraordinarily high binding specificity and affinity between biotin and streptavidin [9–11]. The bond between streptavidin and biotin-conjugated molecules is one of the strongest naturally existing non-covalent bonds, with an equilibrium dissociation constant (KD) between 10-14 and 10-16 M [12,13]. This is stronger than most antibody–antigen interactions, which were reported by the Abcam company and academic investigators to have a KD between 10-6 and 10-9 M or, in the case of very-high-affinity antibodies, a KD of 10-12 M [14–17]. Our approach involves incubating live cells with biotinylated primary antibodies, processing the cells for western blot, and detecting antibody uptake by probing the western blot membrane with either HRP-conjugated or fluorophore-conjugated streptavidin. Biotinylated primary antibodies are available commercially or can be easily generated using biotinylation kits. Additionally, a wide selection of labeled streptavidin is commercially available. Unlike microscopy-based antibody-uptake assays that require image analysis on a cell-by-cell basis, the western blot-based antibody-uptake approach produces population-level results that are quickly quantifiable through mean band intensities normalized against the total protein in the lysates. The expression level of the target integral membrane protein can be cross-correlated with antibody uptake on the same western blot membrane by employing standard western blotting to probe for the integral membrane protein of interest. An additional advantage of our western blot–based approach is that cell lysates and western blot membranes can be preserved for extended periods at -20 °C.
Since this protocol is new and has not been previously validated, we present an extensive Validation of protocol section at the end of this manuscript. Although the Materials and reagents, Procedure, and Data analysis sections for our antibody-uptake assay apply generally to any protein of interest expressed within any cell culture model system, we include specific notes on how we performed the proof-of-principle experiments that are presented in the Validation of protocol section.
Rationale of our proof-of-principle experiments
To demonstrate the utility of this approach, we use a fluorescent protein (FP)-tagged variant of prion protein C179A (PrP*). Untagged PrP*, yellow fluorescent protein (YFP)-PrP*, cerulean fluorescent protein (CFP)-PrP*, and green fluorescent protein (GFP)-PrP* were each shown to leave the ER, traffic to the PM, and rapidly undergo endocytosis [18–22]. The FP-tag, however, does not drive the trafficking of PrP*, but merely provides a detectable label to monitor PrP* trafficking by live-cell imaging [20]. During steady-state conditions, tagged variants of PrP* undergo ER-to-Golgi export via an ER stress-inducible pathway called RESET, which is short for rapid ER stress-induced export [19,20]. After undergoing RESET, PrP* gains access to the PM [18,20]. Since the YFP-tag of YFP-PrP* is derived from and structurally similar to GFP, it is recognized by the same anti-GFP antibodies that bind GFP [20,23]. Critically, YFP-PrP* or GFP-PrP*-expressing cells were previously shown to internalize anti-GFP antibodies based on specific affinity between the anti-GFP antibody and the YFP- or GFP-tag by confocal microscopy and flow cytometry–based approaches [18–21]. In the Validation of protocol section, we present proof-of-principle experiments using anti-GFP antibodies to monitor YFP-PrP* traffic through the PM en route to lysosomes. However, we expect that this antibody-uptake protocol will be applicable to monitor any protein displaying an antigen on an extracellular domain for which there is a biotinylated primary antibody available.
Materials and reagents
Biological materials
1. Healthy cells. This protocol applies to any adherent, proliferating, or differentiated cultured cells.
Note: Since we are monitoring traffic of YFP-PrP* to the cell surface via the RESET pathway for the proof-of-principle experiments presented below in the Validation of protocol section, we opted to use normal rat kidney (NRK) cells. NRK cells were used by Satpute-Krishnan et al. in the Lippincott–Schwartz lab to identify the RESET pathway [20]. In follow-up work, we used them to further dissect the RESET pathway [18,19,22]. These NRK cells were derived from a clonal line previously characterized by the Lippincott-Schwartz lab as adherent cells and used in membrane trafficking studies [24].
Recommendations:
1. Determine the number of replicates per condition within and across experiments.
For experimental results that one plans to quantify, we recommend a minimum of three biological replicates. Each biological replicate should be executed on a different day using freshly prepared medium and drug treatments. For example, in our proof-of-principle experiments, we conducted three biological replicates to obtain measurements and perform statistical analysis (described below). If the experimentalist has little to no experience executing cell culture experiments or carrying out western blots, we recommend a minimum of three technical replicates within a single experiment. This will allow for troubleshooting in the case of varied results.
2. Include a positive control to verify that your biotinylated antibody binds to the antigen (i.e., your integral membrane protein of interest) in the context of the antibody-uptake conditions.
If there is no preexisting evidence that your integral membrane protein of interest traffics to the PM, consider overexpressing a fusion protein to test the antigen–antibody interaction. This fusion protein would include the extracellular domain of your integral membrane protein of interest (i.e., the antigen) on a known PM-resident protein and function as a positive control.
To validate our protocol, we used the following cells for our positive controls:
a. YFP-PrP* NRK cells: These NRK cells stably express the well-characterized RESET substrate, YFP-PrP*, at physiological levels, similar to the prion protein in mouse brain lysate [19,20]. YFP-PrP* includes an N-terminal prolactin signal sequence to ensure ER-translocation, a yellow fluorescent protein (YFP) tag, followed by the Syrian hamster prion protein (PrP) mature domain containing a C179A point mutation [20]. YFP-PrP* and GFP-PrP* were each previously shown to bind and internalize anti-GFP antibodies upon accessing the PM [18–21].
b. YFP-PrP* NRK cells treated with thapsigargin (TG): TG-treatment of YFP-PrP* NRK cells was shown to increase the flux of YFP-PrP* traffic to the PM and antibody uptake when compared to untreated YFP-PrP* NRK cells [20].
3. Include negative controls to determine baseline antibody uptake. This can be cells treated with RNAi or CRISPR knockout technology to remove the integral membrane protein of interest, or cells treated with chemical or siRNA-based inhibitors of the secretory pathway to prevent the integral membrane protein of interest from accessing the PM.
To validate our protocol, we used the following cells for our negative controls:
a. NRK cells: These untransfected cells serve as a negative control for anti-GFP antibody uptake because they do not express an FP-tagged protein. They are the “parental” cell line from which the YFP-PrP* NRK stable cell line was derived.
b. YFP-CD3δ NRK cells: These NRK cells stably express YFP-CD3δ, a well-characterized ER-associated degradation (ERAD) substrate [25,26], at similar levels to the expression of YFP-PrP* in YFP-PrP* NRK cells [19,20]. They serve as a negative control because YFP-CD3δ would not be expected to undergo ER export but is instead degraded at the ER [25,26].
c. YFP-PrP* NRK cells treated with BRD4780: BRD4780-treatment of YFP-PrP* NRK cells was shown to inhibit RESET of YFP-PrP* [18]. Thus, BRD4780 would be expected to block traffic to the PM and thereby prevent anti-GFP antibody uptake.
d. YFP-PrP* NRK cells treated without biotinylated anti-GFP antibody in media: These cells would not be expected to show a streptavidin-binding band by western blot, because no biotinylated antibodies would have been added in the medium for the cells to uptake.
Reagents
Cell culture
1. Cell culture medium
a. Dulbecco’s modified Eagle's medium (DMEM) (Corning, catalog number: 17-205-CV)
b. Fetal bovine serum (FBS) (Corning, catalog number: 35-011-CV)
c. L-glutamine (Corning, catalog number: 25-005-CL)
2. Trypsin (Corning, catalog number: 25-051-CI)
3. Biotinylated primary antibody: biotinylated goat anti-GFP antibody (Genetex, catalog number: GTX26658)
4. Optional: Inducers or inhibitors that promote or block the protein-of-interest’s traffic to the PM. We used the following:
a. Inducer of ER-export via RESET: Thapsigargin (Sigma, catalog number: 586005)
b. Inhibitor of ER-export via RESET: BRD4780, also called AGN1924 hydrochloride (Tocris, catalog number: 1072)
5. Lysosomal degradation inhibitor to prevent degradation of biotinylated antibody and protein of interest: bafilomycin A1 (LC Laboratories, catalog number: B-1080)
6. Wash buffer to wash away excess non-internalized antibody: phosphate-buffered saline (PBS), pH 7.4 (KD Medical, catalog number: RGF-3210)
Western blot
7. 2× Laemmli sample buffer (ready-made 2× sample buffer) (Bio-Rad, catalog number: 1610737)
8. Reducing agent to break disulfide bonds for optimal SDS PAGE and western blot: tris(2-carboxyethyl)phosphine (TCEP) (Sigma, CAS number: 51805-45-9, catalog number: 646547)
9. Protein gels for SDS-PAGE: ready-made 12% Mini-PROTEAN® TGX Stain-FreeTM 15-well gels (Bio-Rad, catalog number: 4568046)
10. Molecular weight ladder: Precision Plus ProteinTM All Blue PreStained Protein Standards (Bio-Rad, catalog number: 1610373)
11. Transfer reagents for western blot: Transfer kit, including PVDF membrane (low fluorescence, 0.45 μm) that requires ethanol for activation, filter paper, and 5× transfer buffer (Bio-Rad, catalog number: 1704275) that requires supplementation with ethanol, 200 Proof (Sigma, catalog number: 459829-2L)
12. Gel running buffer: Tris glycine SDS, pH 8.3 (Santa Cruz Biotechnology, catalog number: sc-296527)
13. Membrane washing buffer: Tris-buffered saline 10×, pH 7.4 (Santa Cruz Biotechnology, catalog number: sc-362308) supplemented with Tween® 20 (Sigma, catalog number: P1379-1L)
14. Blocking buffer: EveryBlot blocking buffer (Bio-Rad, catalog number: 12010020)
15. Labeled streptavidin to detect the internalized biotinylated primary antibody: DyLight800-conjugated streptavidin (Streptavidin-DyLight800, Bio-Rad, catalog number: STAR152D800GA); an alternative is HRP-conjugated streptavidin (Proteintech, catalog number: SA-0000-10)
16. Enhanced chemiluminescence reagents to detect HRP-conjugated streptavidin or secondary antibodies: Chemiluminescence, long duration (Clarity ECL, Bio-Rad, catalog number: 1705061) and chemiluminescence, high sensitivity (Clarity Max ECL, Bio-Rad, catalog number: 1705062)
Solutions
1. Complete DMEM (see Recipes)
2. Thapsigargin (TG) (see Recipes)
3. BRD4780 (also called AGN192403 hydrochloride) (see Recipes)
4. Bafilomycin A1 (see Recipes)
5. Biotinylated goat anti-GFP antibody (see Recipes)
6. 2× Laemmli sample buffer (2× SB) + tris(2-carboxyethyl)phosphine (TCEP) (see Recipes)
7. Tris glycine SDS (see Recipes)
8. TBST (see Recipes)
9. Streptavidin-DyLight800 (see Recipes)
Recipes
1. Complete DMEM
10% FBS and 2 mM L-glutamine in DMEM
2. Thapsigargin (TG)
0.1 μM TG in complete DMEM
3. BRD4780 (also called AGN192403 hydrochloride)
100 μM in complete DMEM
4. Bafilomycin A1
500 nM bafilomycin A1 in complete DMEM
5. Biotinylated goat anti-GFP antibody
1 μg/mL in complete DMEM
6. 2× Laemmli sample buffer (2× SB) + tris(2-carboxyethyl)phosphine (TCEP)
5 mM TCEP in 2× SB
7. Tris Glycine SDS
0.025 M Tris, 0.192 M glycine, and 0.1% SDS at pH 8.3
8. TBST
250 mM Tris, 27 mM KCl, 1.37 M NaCl at pH 7.4 with 0.1% Tween® 20
9. Streptavidin-DyLight800
Diluted 1:5,000 in EveryBlot Bio-Rad blocking buffer
Laboratory supplies
1. 10 cm tissue culture-treated dish (VWR, catalog number: 25382-428)
2. 6-well tissue culture-treated plates, sterile, flat-bottom wells (Genesee, catalog number: 25-105MP)
3. Gel loading tips, 1–200 μL pipette tip for gel loading (Bio-Rad, catalog number: 12021140)
4. Boil-proof microfuge tubes (Genesee Scientific, catalog number: 24-282C)
Equipment
1. Gel electrophoresis apparatus (Mini-PROTEAN® Tetra Electrode Assembly) (Bio-Rad, catalog number: 1658037)
2. Western blot transfer apparatus (Trans-Blot Turbo Transfer System) (Bio-Rad, catalog number: 1704150)
3. Gel and western blot documentation apparatus (ChemiDocTM MP Imaging System) (Bio-Rad, catalog number: 12003154)
Software and datasets
1. FIJI “Fiji is just ImageJ” (National Institutes of Health)
2. Excel (Microsoft)
3. Prism (GraphPad)
Procedure
文章信息
稿件历史记录
提交日期: Sep 4, 2025
接收日期: Oct 8, 2025
在线发布日期: Oct 29, 2025
出版日期: Nov 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Graninger, A. and Satpute-Krishnan, P. (2025). Monitoring Endocytosis of Integral Membrane Proteins Using Western Blot-Based Detection of Biotinylated Antibody Uptake. Bio-protocol 15(22): e5511. DOI: 10.21769/BioProtoc.5511.
分类
细胞生物学 > 基于细胞的分析方法 > 蛋白质分泌
细胞生物学 > 基于细胞的分析方法 > 转运
生物化学 > 蛋白质 > 免疫检测 > 免疫印迹法(WB )
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










