- EN - English
- CN - 中文
Library Preparation for Genome-Wide DNA Methylation Profiling
全基因组DNA甲基化分析的文库构建方法
发布: 2025年11月05日第15卷第21期 DOI: 10.21769/BioProtoc.5488 浏览次数: 1587
评审: Migla MiskinyteAnonymous reviewer(s)
Abstract
DNA methylation is a fundamental epigenetic mark with critical roles in epigenetic regulation, development, and genome stability across diverse organisms. Whole genome bisulfite sequencing (WGBS) enables single-base resolution mapping of cytosine methylation patterns and has become a standard method in epigenomics. This protocol provides a detailed, step-by-step workflow for WGBS library construction starting from genomic DNA. It includes steps of RNaseA treatment, DNA shearing, end-repair and A-tailing, adapter ligation, bisulfite conversion, library amplification, and quantification. Notably, the method uses self-prepared reagents and customizable index systems, avoiding the constraints of commercial library preparation kits. This flexibility supports cost-effective, scalable methylome profiling, suitable for diverse experimental designs, including high-throughput multiplexed sequencing.
Key features
• Provides a comprehensive workflow for whole-genome bisulfite sequencing (WGBS) library preparation without relying on commercial kits.
• Enables flexible multiplexing by allowing users to customize index systems during oligonucleotide synthesis.
• Offers high-resolution, unbiased DNA methylation profiling at single-base resolution.
• Optimized for cost-effective and scalable applications, including high-throughput sample processing.
• Compatible with a wide range of DNA inputs and adaptable to different species and experimental designs.
Keywords: DNA methylation (DNA甲基化)Graphical overview
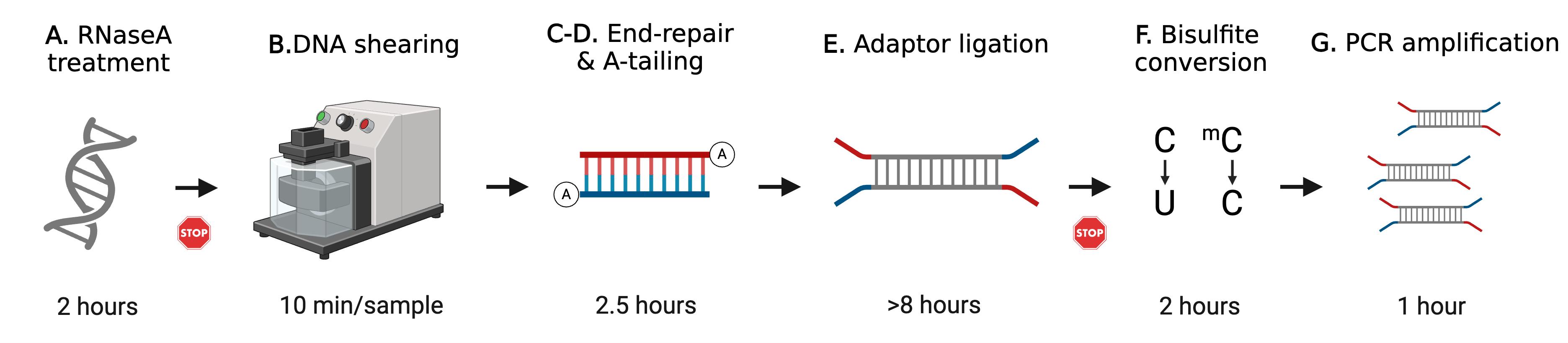
Schematic overview of whole genome bisulfite sequencing (WGBS) library preparation. This protocol describes step-by-step WGBS library preparation. Starting from genomic DNA (gDNA), RNaseA treatment removes excessive RNA. gDNA is then fragmented by ultrasonication. End-repair and A-tailing allow DNA fragment ends to be modified for adapter ligation. After a few cycles of PCR amplification, the final WGBS library is then quantified and verified for concentration and size distribution. The STOP signs represent safe stop points.
Background
DNA methylation is one of the most studied epigenetic modifications, involving the addition of a methyl group to the fifth carbon of a cytosine (5C), resulting in 5-methylcytosine (5mC). Under high temperature for a period, the double-stranded DNA is denatured, sodium bisulfite is able to react with the unmethylated cytosines, and a sulfonated cytosine intermediate is formed. This intermediate is then converted through hydrolytic deamination, followed by an alkaline treatment, and the cytosine is fully converted to uracil (U). This process, called bisulfite conversion, is the basis of bisulfite sequencing (BS-seq), which has been developed to profile DNA methylation systematically when coupled to next-generation sequencing [1].
Whole genome bisulfite sequencing (WGBS) is one of the most powerful tools to profile genome-wide DNA methylation, capturing most cytosines in each context, e.g., CG, CHG, and CHH, without bias [1,2]. WGBS library construction usually takes 3–5 days. Several commercial WGBS library preparation kits have been released for different applications, e.g., ultralow input or short bisulfite conversion time. These kits provide all-in-one packages that include bisulfite conversion and library construction reagents. Though convenient, the commercial kits usually come with defined indexes and reaction numbers that can lead to limitations with respect to multiplexing with other sequencing libraries and higher costs. In this protocol, we describe WGBS library construction steps and provide a flexible alternative for researchers to generate a WGBS library without library preparation kits. The presented protocol has been applied to rice [3] and Arabidopsis [4,5], and is assumed to be compatible with other species.
Materials and reagents
Biological materials
1. Genomic DNA (gDNA) from the organism of interest; 500 ng or more gDNA is recommended for this protocol
2. Phage lambda DNA (Thermo Scientific, catalog number: SD0021)
Reagents
1. DNAZapTM PCR DNA degradation solutions (Invitrogen, catalog number: AM9890 or equivalent); store at 4 °C
2. RNase A, DNase and protease-free (Thermo Scientific, catalog number: EN0531); store at -20 °C
3. AMPure XP beads (Beckman Coulter, catalog numbers: A63880, A63881, or A63882); store at 4 °C
4. Molecular biology–grade absolute ethanol (Fisher Scientific, catalog number: BP2818110); store at room temperature (RT)
5. UltraPure DNase/RNase-free distilled water (Invitrogen, catalog number: 10977015); store at RT
6. UltraPure 1 M Tris-HCl, pH 8.0 (Invitrogen, catalog number: 15568025); store at RT
7. Klenow Fragment 3′→5′ exonuclease (New England Biolabs, catalog number: M210S); store at -20 °C
8. T4 DNA ligase (New England Biolabs, catalog number: M0202S); store at -20 °C
9. EpiTect Fast Bisulfite Conversion kit (Qiagen, catalog number: 59802); store parts at different temperatures according to the manufacturer’s guide
10. MinElute PCR Purification kit (Qiagen, catalog number: 28004); store the spin columns at 4 °C and the remaining at RT
11. PfuTurbo Cx hotstart DNA polymerase (Agilent Technologies, catalog number: 600410); store at -20 °C
12. Deoxynucleotide (dNTP) solution mix (New England Biolabs, catalog number: N0447S); store at -20 °C
13. dATP solution (New England Biolabs, catalog number: N0440S); store at -20 °C
14. Qubit dsDNA BR Quantification Assay kit (Invitrogen, catalog number: Q32850); store the DNA standard at 4 °C and the remaining at RT; keep the dye reagent from light
15. TapeStation D1000 ScreenTape (Agilent Technologies, catalog number: 5067-5582); store at 4 °C
16. TapeStation D1000 ladder (Agilent Technologies, catalog number: 5067-5586); store at 4 °C
17. TapeStation D1000 sample buffer (Agilent Technologies, catalog number: 5067-5602); store at 4 °C
Solutions
1. Elution buffer (EB) (see Recipes)
2. RNaseA stock (see Recipes)
3. RNaseA reaction (see Recipes)
4. End-repair mix (see Recipes)
5. A-tailing mix (see Recipes)
6. Ligation mix (see Recipes)
7. Amplification mix (see Recipes)
8. Qubit working solution (see Recipes)
9. 80% ethanol (EtOH) (see Recipes)
Recipes
1. Elution buffer (EB)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| UltraPure 1 M Tris-HCl, pH 8.0 | 10 mM | 10 μL |
| UltraPure DNase/RNase-free distilled water | n/a | 990 μL |
| Total | 10 mM | 1 mL |
Caution: Store at RT.
2. RNaseA stock
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| RNase A, DNase and protease-free | 10 mg/mL | 10 mg |
| UltraPure DNase/RNase-free distilled water | n/a | 1 mL |
| Total | 10 mg/mL | 1 mL |
Caution: Aliquot 10 μL per tube and store at -20 °C. Thaw on ice before use. Do not freeze and thaw.
3. RNaseA reaction
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| RNase A stock | 100 μg/mL | 0.5 μL |
| gDNA solution | n/a | n/a |
| UltraPure DNase/RNase-free distilled water | n/a | n/a |
| Total | 100 μg/mL | 50 μL |
Caution: Do not re-freeze the RNAseA stock.
4. End-repair mix
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sheared DNA | n/a | 30 μL |
| NEBNext end repair reaction buffer (10×) | 1× | 5 μL |
| NEBNext end repair enzyme mix | n/a | 2.5 μL |
| UltraPure DNase/RNase-free distilled water | n/a | 12.5 μL |
| Total | n/a | 50 μL |
5. A-tailing mix
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| End-repaired DNA | n/a | 30 μL |
| 10× NEB buffer 2 | 1× | 5 μL |
| 1mM dATP | 0.2 mM | 10 μL |
| 3′→5′ Klenow exonuclease | n/a | 3 μL |
| UltraPure DNase/RNase-free distilled water | n/a | 2 μL |
| Total | n/a | 50 μL |
6. Ligation mix
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 10× T4 DNA ligation buffer | 1× | 3 μL |
| Adapter | 2 μM | 6 μL |
| A-tailed DNA | n/a | 20 μL |
| T4 DNA Ligase | n/a | 1 μL |
| Total | n/a | 30 μL |
7. Amplification mix
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Forward primer | 0.5 μM | 2.5 μL |
| Reverse primer | 0.5 μM | 2.5 μL |
| 10mM dNTP | 0.2 mM | 1 μL |
| 10× Pfu Turbo Cx reaction buffer | 1× | 5 μL |
| Pfu Turbo Cx hotstart DNA polymerase | n/a | 1 μL |
| UltraPure DNase/RNase-free distilled water | n/a | 22 μL |
| Bisulfite converted DNA | n/a | 16 μL |
| Total | n/a | 50 μL |
8. Qubit working solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Qubit reagent | n/a | 1 μL |
| Qubit buffer | n/a | 199 μL |
| Total | n/a | 200 μL |
9. 80% ethanol (EtOH)
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Absolute ethanol | n/a | 4 mL |
| UltraPure DNase/RNase-free distilled water | n/a | 1 mL |
| Total | n/a | 5 mL |
Laboratory supplies
1. Low-retention barrier tips (Neptune, catalog numbers: 63300746, 63300757, 63300759, and 63300749, or equivalent)
2. microTUBE AFA fiber pre-slit snap-cap 6 × 16 mm (Covaris, catalog number: 520045)
3. 1.5 mL EpiTube (Eppendorf, catalog number: 022431021)
4. PCR strips (USA Scientific, catalog number: 1402-4700, or equivalent)
5. Qubit assay tube (Axygen, catalog number: PCR-05-C)
6. TapeStation tips (Agilent Technologies, catalog number: 5067-5099)
7. TapeStation tube strips (Agilent Technologies, catalog number: 401428)
8. TapeStation tube strip caps (Agilent Technologies, catalog number: 401425)
Equipment
1. Ultrasonicator (Covaris, model: S220)
2. TapeStation (Agilent Technologies, model: 4200, or equivalent)
3. Qubit fluorometer (Invitrogen, model: 4.0, or equivalent)
4. Thermocycler (Bio-Rad, model: T100, or equivalent)
5. Microcentrifuge (Eppendorf, model: 5425, or equivalent)
6. ThermoMixer (Eppendorf, model: R, or equivalent)
7. Vortexer (Scientific Industries, model: Vortex-Genie 2, or equivalent)
8. DynaMag2 (Invitrogen, catalog number: 12321D)
9. DynaMag-PCR magnet (Invitrogen, catalog number: 492025)
10. MS3 vortexer (IKA, catalog number: 0003617000)
11. NanoDrop (Thermo Scientific, model: ND-1000, or equivalent)
Procedure
文章信息
稿件历史记录
提交日期: Jul 24, 2025
接收日期: Sep 24, 2025
在线发布日期: Oct 16, 2025
出版日期: Nov 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Hsu, F., Pellegrini, M. and Chen, P. Y. (2025). Library Preparation for Genome-Wide DNA Methylation Profiling. Bio-protocol 15(21): e5488. DOI: 10.21769/BioProtoc.5488.
分类
分子生物学 > DNA > DNA 测序
分子生物学 > DNA > DNA 修饰
系统生物学 > 表观基因组学 > DNA 甲基化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













