- EN - English
- CN - 中文
SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
基于SunTag系统的秀丽隐杆线虫单分子翻译成像
发布: 2025年10月20日第15卷第20期 DOI: 10.21769/BioProtoc.5486 浏览次数: 2260
评审: Munenori IshibashiAnonymous reviewer(s)
Abstract
Translation is a key step in decoding the genetic information stored in DNA. Regulation of translation is an important step in gene expression control and is essential for healthy organismal development and behavior. Despite the importance of translation regulation, its impact and dynamics remain only partially understood. One reason is the lack of methods that enable the real-time visualization of translation in the context of multicellular organisms. To overcome this critical gap, microscopy-based methods that allow visualization of translation on single mRNAs in living cells and animals have been developed. A powerful approach is the SunTag system, which enables real-time imaging of nascent peptide synthesis with high spatial and temporal resolution. This protocol describes the implementation and use of the SunTag translation imaging system in the small round worm Caenorhabditis elegans. The protocol provides details on how to design, carry out, and interpret experiments to image translation dynamics of an mRNA of interest in a cell type of choice of living C. elegans. The ability to image translation live enables better understanding of translation and reveals the mechanisms underlying the dynamics of cell type–specific and subcellular localization of translation in development.
Key features
• Describes how to visualize translation of single mRNA molecules in living C. elegans.
• Can be used to visualize translation in space and time in C. elegans larvae and embryos.
Keywords: mRNA translation (mRNA翻译)Graphical overview
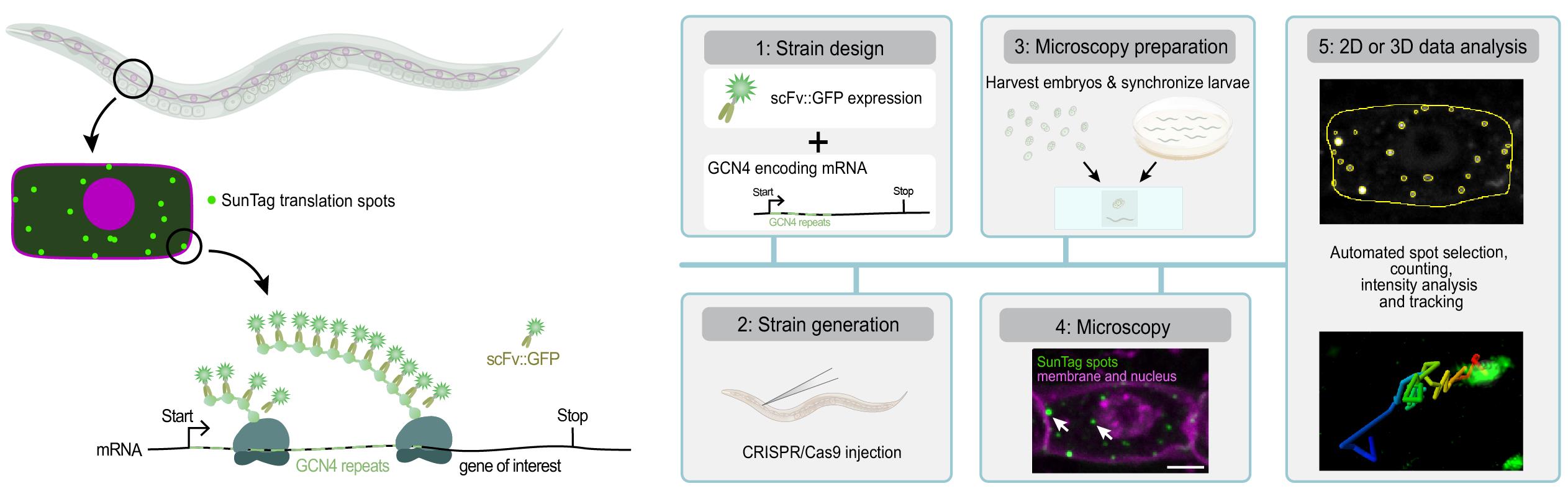
Visualizing translation in C. elegans using the SunTag system. The SunTag system enables real-time imaging of translation in specific cells during C. elegans development, from embryos to larvae. To visualize translation, a gene of interest is fused to an array of SunTag (GCN4) peptides. The GCN4 peptides emerge first from the ribosome and are bound by the co-expressed, GFP-labeled SunTag antibody (scFv::GFP), resulting in a bright fluorescent spot at the site of translation and thus allowing real-time observation of protein synthesis. This protocol provides step-by-step instructions for designing and generating strains, preparing animals for microscopy, performing live translation imaging, and analyzing data in 2D or 3D.
Background
Translation of mRNAs into proteins is a key step in gene expression, and its regulation is essential for cellular functioning. Translation is fast, reversible, and allows for post-transcriptional control of gene expression, especially when transcription is silenced. For example, during early embryogenesis, translation regulation of maternally provided mRNAs directs lineage specification and embryonic development [1]. In addition, translation regulation enables localized protein synthesis, for example, in highly polarized cells such as neurons [2]. Due to its vital role in tissue homeostasis [3], defects in either translation efficiency or fidelity affect healthy aging [4] and underlie many human diseases, including neurological disorders and cancer [5,6].
In recent years, different methods have provided many new and important insights into the regulation of translation, yet many questions remain. For instance, although translational heterogeneity between individual mRNAs is increasingly recognized [7], it is still unclear how widespread this heterogeneity is, under what conditions it occurs, and how it is regulated in space and time—particularly in multicellular organisms. Addressing these questions requires tools that reveal when and where specific mRNAs are translated. A major limitation of many current methods is their dependence on population-level measurements and the requirement for fixation or lysis, capturing only static snapshots and average translation efficiencies of thousands of mRNAs. For example, ribosome profiling [8,9] is a powerful sequencing-based method to assess global translation, but lacks spatial and temporal resolution. Newer methods, such as RIBOmap [10], partially address this by mapping the location of translated mRNAs in tissues, but offer limited insight into translation efficiencies of individual mRNAs or in changes over time. To overcome these limitations, live-imaging of translation has been developed, using the SunTag system [11–16], which enables real-time analysis of the location, timing, and efficiency of translation of specific mRNAs in living cells and animals. Here, we describe how to use the SunTag system in C. elegans.
The SunTag system relies on two components: 1) an mRNA of interest encoding an array of SunTag peptides (GCN4s), and 2) an anti-GCN4 SunTag antibody labeled with superfolder GFP (scFv::GFP). During mRNA translation, the GCN4 peptides are synthesized and rapidly bound by the co-expressed scFv::GFP antibody (Figure 1A). As multiple ribosomes simultaneously translate the mRNA and each ribosome produces multiple GCN4 repeats, the scFv::GFP accumulates at the site of translation, which appears as a bright fluorescent spot under the microscope (Figure 1 and Video 1). The intensity of each spot is determined by the number of ribosomes that translate the mRNA. Together, the number of translation spots and the intensity of the spots provide a quantitative measure of translation efficiency. Moreover, tracking individual SunTag spots over time yields detailed insight into the spatial and temporal dynamics of translation.
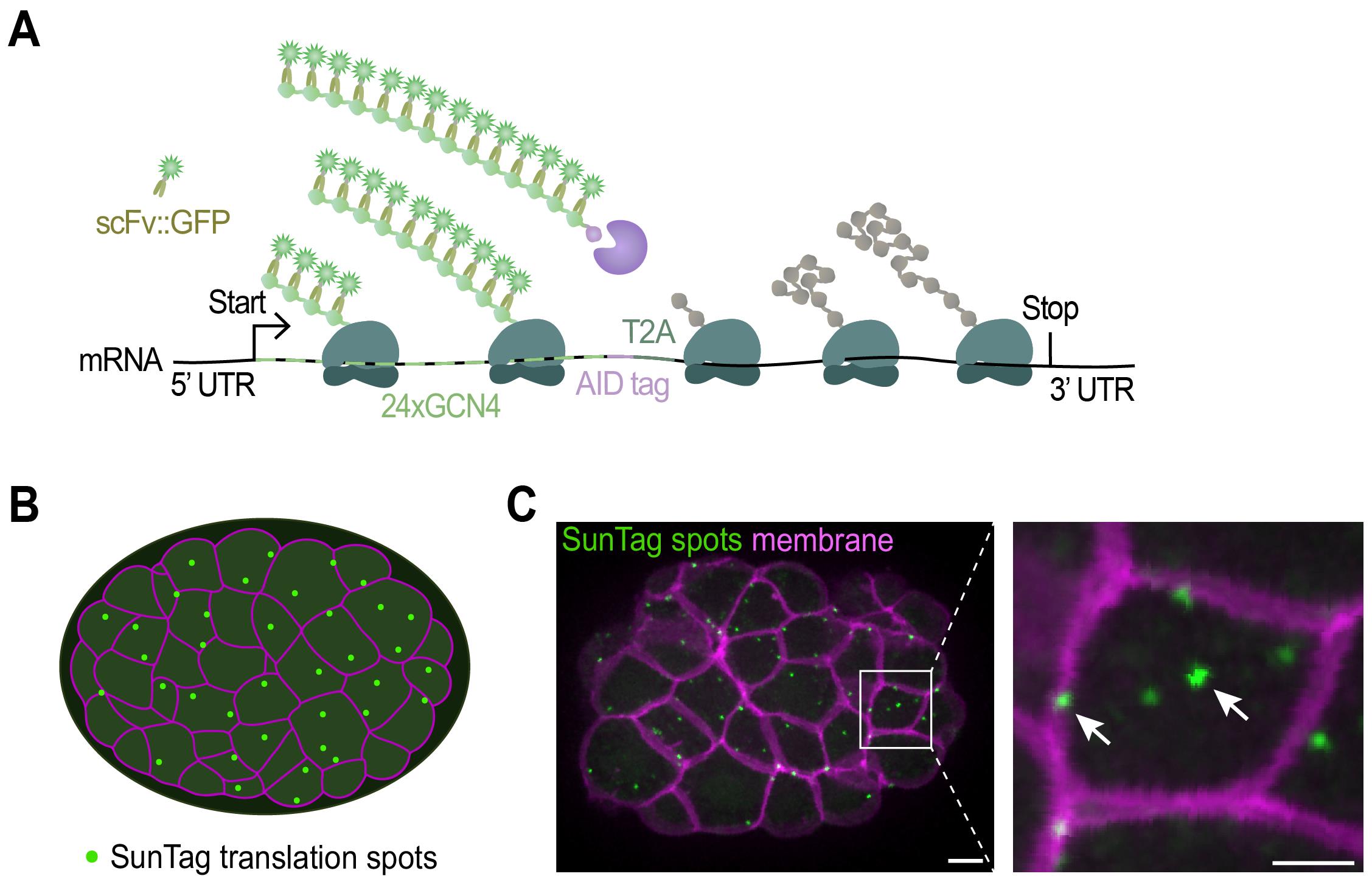
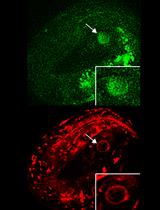
Figure 1. Imaging translation of single mRNA molecules. (A) Schematic representation of the fluorescent labeling of nascent polypeptides using the SunTag system. When the mRNA is translated, scFv::GFP antibodies will bind to the SunTag peptides that emerge from the ribosome, resulting in a bright fluorescent spot at the site of translation and thus allowing real-time observation of protein synthesis. (B) Schematic representation of an embryo with membranes in magenta and translation spots in green. (C) Representative image of an embryo expressing scFv::GFP and the translational reporter (eft-3p::24xGCN4::T2A::BFP::h2b::tbb-2 3’UTR). Translation is shown in green, membranes in magenta. A zoomed-in view of the white-boxed area is shown to the right. Scale bars, 5 μm (left panel) or 2.5 μm (right panel).
Materials and reagents
Biological materials
1. OP50 Escherichia coli (OP50) (C. elegans Genetic Center)
2. eft-3p::Cas9 plasmid (50 ng/μL) (Addgene, #46168)
3. Single-guide RNA (sgRNA) plasmid (50 ng/μL) (IDT)
4. pRF4 [rol-6(su1006)] (40 ng/μL) [22]
5. Repair template plasmid (50 ng/μL) (Addgene, # depends on specific aim)
6. Co-injection marker plasmid (2.5 ng/μL) (Addgene, # depends on specific aim)
7. pcDNA4TO-24xGCN4_v4-kif18b-24xPP7 (Addgene, #74928) [12]
8. Useful strains for SunTag imaging (see Tables 1 and S1 and Figure 2)
Table 1. Available SunTag strains
| Strain name | Genotype See Table S1 for a description of how the strains were generated, including the sequence of the constructs, and Figure 2 for a graphical representation of the strains |
| SUR10 | cxTi10816(rubSi7[eft-3p::24xGCN4::t2a::tagbfp::h2b::tbb-2 3’UTR) IV; ttTi5605(rubSi6[eft-3p::scfv(glo)::gfp(smu-1 introns)::tbb-2 3’UTR II |
| SUR54 | cxTi10816(rubSi8[eft-3p::24xGCN4::AID::t2a::tagbfp::h2b::tbb-2 3’UTR *rubSi7]) IV; ttTi5605(rubSi6[eft-3p::scfv(glo)::gfp(smu-1 introns)::tbb-2 3’UTR;]) II; ltIs44[pie-1p::mCherry::PH(PLC1delta1) + unc-119(+)]; wrdSi23[eft-3p::TIR1::F2A::mTagBFP2::AID*::NLS::tbb-2 3'UTR] (I:-5.32) |
| SUR72 | ttTi5605(rubSi6[eft-3p::scfv(glo)::gfp(smu-1 introns)::tbb-2 3’UTR;]) II |
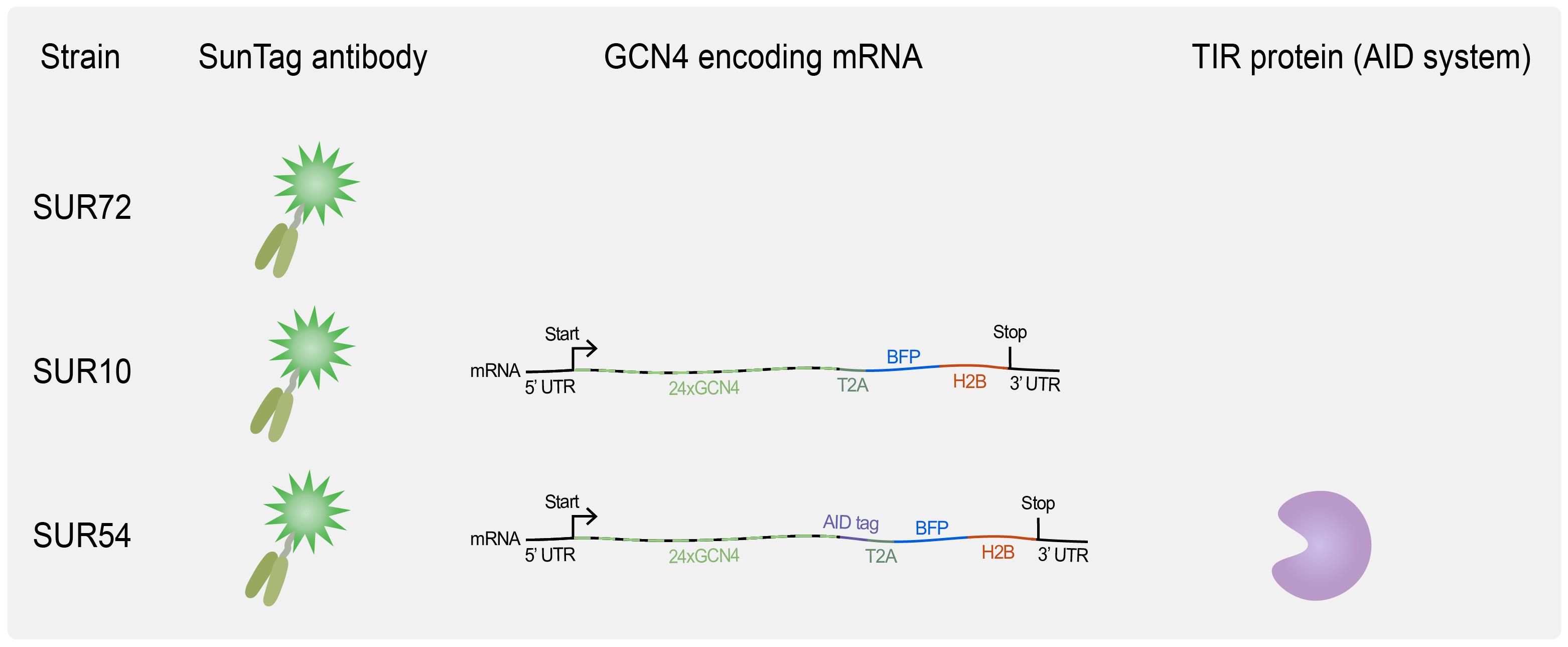
Figure 2. Overview of available C. elegans strains for translation imaging. SUR72 expresses the SunTag antibody scFv::GFP, SUR10 combines scFv::GFP expression with the expression of a general translation reporter, and SUR54 adds the auxin-mediated degradation system, which requires TIR1 expression (see Biological materials 8 for genotypes).
9. Useful plasmids (see Tables 2 and S2)
Table 2. Useful plasmids for implementing the SunTag system
| Plasmid | Description of the most important sequences present in the plasmid. Please note that the full sequence is provided in Table S2 | Addgene ID |
| pSRG_11 | SunTag antibody | #245121 |
| pSRG_12 | SunTag reporter without AID system | #245120 |
| pSRG_100 | 3xGCN4 | #246457 |
| pSRG_101 | KasI::24xGCN4::T2A::AvrII | #246458 |
| pJJS_001 | mIAA7::GFP::Linker | #188324 |
Reagents
1. Cas9 protein (0.25 μg/μL) (IDT, catalog number: 1081058)
2. TracRNA (0.1 μg/μL) (IDT, catalog number: 1073190)
3. Locus-specific crRNA can be ordered from IDT, sequence dependent on the gene of interest (56 ng/μL) (IDT)
4. Primers containing 5′ SP9 modifications can be ordered from IDT, sequence dependent on the gene of interest (IDT)
5. ssODN can be ordered from IDT, sequence dependent on the gene of interest (0.11 μg/μL) (IDT), see [23] for how to design
6. NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, catalog number: 740609.250)
7. 1 mM auxin (IAA) (Alfa Aesar, CAS number: 87-51-4)
8. 10 mM Tetramisole hydrochloride solution (Sigma, CAS number: 5086-74-8)
9. Q5® High-Fidelity PCR Kit (New England Biolabs, catalog number: E0555)
10. UltraPure agarose (Invitrogen, catalog number: 16500500)
11. NaCl (Merck, CAS number: 7647-14-5)
12. Na2HPO4·2H2O (Merck, CAS number: 10028-24-7)
13. KH2PO4 (Merck, CAS number: 7778-77-0)
14. NaOH pellets (Merck, CAS number: 1310-73-2)
15. Agar powder (Boom, CAS number: 9002-18-0)
16. BactoTM peptone (Gibco, catalog number: 211677)
17. K2HPO4 (Carl Roth, CAS number: 7758-11-4)
18. MgSO4·7H2O (Sigma, CAS number: 10034-99-8)
19. CaCl2·2H2O (Sigma, CAS number: 10035-04-8)
20. Cholesterol (Sigma, CAS number: 57-88-5)
21. Ethanol (Merck, CAS number: 64-17-5)
22. TWEEN® 20 (Sigma-Aldrich, CAS number: 9005-64-5)
23. Commercial thin bleach (<5% NaOCl) (C.I.V. Superunie B.A., catalog number: 8710624315320)
24. Useful PCR primers, see Table 3
Table 3. Useful primers for genotyping SunTag strains
| Purpose | Primer sequence | Target sequence |
| Genotype SunTag antibody1 | (F) CTGATTGACCCGATGAAATACGGATG | tti5605 MosSci site, Chr II, flanking SunTag antibody |
| (R) CTCACTCGTTTAGGCTATTCCCC | tti5605 MosSci site, Chr II, flanking SunTag antibody | |
| Genotype SunTag reporter2 | (F) GGAAGTCGGAGTACAAATTCG | cxTi10816 MosSci site, Chr IV, flanking SunTag reporter |
| (R) CGAACCATTCTTCCTGGC | cxTi10816 MosSci site, Chr IV, flanking SunTag reporter | |
| Genotype SunTag antibody or reporter3 | (F) AGTTGACAATAAAAGACCAAAGGTGC | eft-3promoter, within SunTag antibody locus and within SunTag reporter locus |
| Genotype TIR1 | (F) TGTCGACCGCTAGTGTAGCTTAC | Genomic Homology L ttTi4348, Chr I |
| (R) CGTCTCTCCACGATTTACACACTATTTG | Genomic Homology R ttTi4348, Chr 1 | |
| (R) CTGGCCGTCGTTTTACAGGA | let-858 UTR | |
| Amplify 24xGCN4::T2A from pSRG_12 for CRISPR/Cas-9 | (F) [N]30CATGGGCGCCATCATACACGCGGCCGCAATCAT | 5′ end of 24xGCN4::T2A |
| (R) [N]30TGGTCCTGGGTTCTCCTCGACGTC | 3′ end of 24xGCN4::T2A | |
| Amplify 3xGCN4 from pSRG_100 for CRISPR/Cas KI | (F) [N]30ATGGAGGAATTATTGAGCAAGAACTACCAC | 5′ end of 3xGCN4::linker |
| (R) [N]30AGAGCCGGATCCCTTCTTGAGAC | 3′ end of 3xGCN4::linker | |
| Amplify AID from Addgene # 188324 for CRISPR/Cas KI | (F) [N]30GGATTCTCCGAGACCGTCGA | 5′ end of miniIAA7 |
| (R) [N]30GGAGGAGGTCTTTTGTTGGGTC | 3′ end of miniIAA7 |
2Combine with 3 for genotyping the SunTag reporter
Solutions
1. Nematode growth medium (NGM) (see Recipes)
2. NGM with 1 mM auxin (NGM + IAA) (see Recipes)
3. M9 buffer (see Recipes)
4. Bleach solution (see Recipes)
5. KPO4 buffer, pH 6 (see Recipes)
6. PCR reaction master mix (see Recipes)
Recipes
1. Nematode growth medium (NGM)
| Reagent | Final concentration | Quantity or volume for 1 L |
| NaCl | 50 mM | 3 g |
| BactoTM peptone | 2.5 g/L | 2.5 g |
| Agar powder | 17 g/L | 17 g |
| Deionized H2O | n/a | 975 mL |
| Autoclave (20 min, 121 °C), cool until 55 °C, and add the following | ||
| 5 mg/mL cholesterol, diluted in 100% ethanol | 0.005 g/L | 1 mL |
| 1 M CaCl2 solution | 1 mM | 1 mL |
| 1 M MgSO4 solution | 1 mM | 1 mL |
| 1 M KPO4 buffer, pH 6 (see Recipe 5) | 25 mM | 25 mL |
2. NGM with 1 mM Auxin (NGM + IAA)
Prepare in the same way as NGM (see Recipe 1). After the autoclaving step, also include 1 mL of 1 M IAA (diluted in 100% ethanol).
| Reagent | Final concentration | Quantity or volume for 1 L |
|---|---|---|
| 1 M IAA (indole-3-acetic acid), diluted in 100% ethanol | 1 mM | 1 mL |
Critical: Always prepare the 1 M IAA solution fresh before adding it to the medium. IAA precipitates fast.
Critical: Protect IAA from light.
3. M9 buffer
| Reagent | Final concentration | Quantity or volume for 1 L |
| KH2PO4 | 22 mM | 3 g |
| Na2HPO4·2H2O | 42 mM | 7.48 g (6 g anhydrous) |
| NaCl | 85.5 mM | 5 g |
| Deionized H2O | n/a | to 1 L |
| Autoclave (20 min, 121 °C), cool until 55 °C, and add the following | ||
| 1 M MgSO4 solution | 1 mM | 1 mL |
4. Bleach solution
| Reagent | Final concentration | Volume for bleaching one 15 mL tube of hermaphrodites |
|---|---|---|
| Commercial thin bleach (<5% NaOCl) | <1.6% | 1.6 mL |
| 10 M NaOH | 251 mM | 125 μL |
| Deionized H2O | n/a | 3.26 mL |
| Total | n/a | 4.985 mL |
5. KPO4 buffer, pH 6
| Reagent | Final concentration | Quantity or volume for 1 L |
|---|---|---|
| KH2PO4 | 0.8 M | 108.3 g |
| K2HPO4 | 0.2 M | 35.6 g |
| Deionized H2O | n/a | to 1 L |
6. PCR reaction master mix
| Reagent | Final concentration | Volume for 50 μL reaction |
|---|---|---|
| 5× Q5 reaction buffer | 1× | 10 μL |
| 10 mM dNTPs | 200 μM | 1 μL |
| 10 μM forward primer | 0.5 μM | 2.5 μL |
| 10 μM reverse primer | 0.5 μM | 2.5 μL |
| Template DNA | 0.5–500 ng | variable |
| Q5 High-Fidelity DNA polymerase | 0.02 U/μL | 0.5 μL |
| 5× Q5 high GC enhancer | 1× | 10 μL |
| Nuclease-free water | to 50 μL |
Laboratory supplies
1. Tubes, 15 mL, uncoated (Sarstedt, catalog number: 62.554.502)
2. Worm pick, custom-made from 99.95% platinum wire (ø 0.3 mm, 1.55 g/m) attached to a glass Pasteur pipette
3. Pipette tips [Greiner, Sapphire, catalog number: 771352 (10 μL), 775352 (200 μL), or 777352 (100 μL)]
4. 5 mL serological pipettes (Sarstedt, catalog number: 86.1253.001)
5. 10 mL serological pipettes (Sarstedt, catalog number: 86.1254.001)
6. Petri dishes 60 × 15 mm (Corning, catalog number: BP53-06)
7. Culture dish, tissue culture treated, 60 mm for larvae synchronization (Cellstar®, catalog number: 628160)
8. 1.5 mL Eppendorf tubes (Eppendorf, catalog number: 0030108442)
9. Needles (for splaying worms) 26 gauge (other gauge needles work as well) (Terumo, catalog number: AN*2613R1)
10. Nitrile gloves (for gel clean-up and getting older embryos) (Avantor, catalog number: 112-2372)
11. Microscope slides (Avantor, catalog number: 631-1552)
12. Borosilicate glass coverslips, thickness No. 1 (Carl Roth, catalog number: H875.2)
13. Lead-free steam indicator tape (3MTM ComplyTM, model: 1322-12MM)
14. Thin wall glass capillaries (WPI, catalog number: TW100F-6)
15. Microloader 100 mm pipette tips 0.5–20 μL (Calibre Scientific, catalog number: 5242956003)
16. 20 μm Nylon net filters (Millipore, catalog number: NY2004700)
17. Plastic funnel (Corning, catalog number: 610P-30)
Equipment
1. Spinning disk confocal microscope [Nikon, model: Nikon Eclipse Ti2-E (motorized) with perfect focus]
2. 100× oil immersion objective (Nikon, model: Nikon CFI Plan Apo λ 100×/1.45 0.13 mm WD OIL 71)
3. Detector (Teledyne Vision Solutions, model: 01-PRIME-BSI-R-M-16-C)
4. Eppendorf Centrifuge 5810 R (Eppendorf, catalog number: 5811000015)
5. Dissection stereomicroscope (Leica, catalog number: 10450814)
6. Pipettes (Eppendorf, catalog number: 2231001169)
7. RF3000 Pipette Controller (Heathrow Scientific HS3000, catalog number: Z767670)
8. Incubator (VDW Cool Systems, model: CI 625 W PRO PID)
9. Injection microscope (Zeiss, model: Axio Observer.A1)
10. Microinjector (Eppendorf, model: FemtoJet 4x)
11. Micromanipulator (Eppendorf, model: TransferMan 4r)
12. Needle puller (pre-made injection needles can also be used) (Sutter Instrument Co., catalog number: P-2000)
13. Thermo ScientificTM OwlTM EasyCastTM B1A Mini Gel Electrophoresis System (Thermo Scientific, catalog number: B1A-BP)
Software and datasets
1. ImageJ (NIH, https://imagej.net/ij/download.html) or "FIJI Is Just ImageJ" FIJI (NIH, http://fiji.sc/Fiji)
2. ComDet v.0.5.5. All data and code have been deposited to GitHub: https://github.com/ekatrukha/ComDet/tree/0.5.5 (access date, 05/13/2022) [23]
3. Imaris File Converter (Bitplane, Belfast, UK, v. 10.0.1, December 2022)
4. Imaris software (Bitplane, Belfast, UK, v. 10.0.1, December 2022), including Essentials and Tracking packages
5. Microsoft Excel (Microsoft, Redmon, WA, Office 365, December 2022)
Procedure
文章信息
稿件历史记录
提交日期: Jul 22, 2025
接收日期: Sep 16, 2025
在线发布日期: Oct 13, 2025
出版日期: Oct 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
van der Salm, E., Kontogiannis, S. and Ruijtenberg, S. (2025). SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans. Bio-protocol 15(20): e5486. DOI: 10.21769/BioProtoc.5486.
分类
分子生物学 > RNA > mRNA 转译
细胞生物学 > 细胞成像 > 活细胞成像
发育生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link