- EN - English
- CN - 中文
Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Cluster FLISA——用于比较不同细胞系蛋白表达效率及蛋白亚基聚集状态的方法
(*contributed equally to this work) 发布: 2025年11月05日第15卷第21期 DOI: 10.21769/BioProtoc.5484 浏览次数: 1209
评审: Gunter MaubachSupreet BhattacharyaAnonymous reviewer(s)
Abstract
Nowadays, recombinant proteins are the focus of various research fields, and their use ranges from therapeutic investigations to cellular model systems for the development of therapeutic approaches. Cell systems used for the expression of recombinant proteins should be comparable in terms of yield and expression efficiency. In many research fields, it is desirable to obtain high protein concentrations. A method that combines an easy workflow with rapid results and affordable costs remains missing, and a standardized approach to determining protein concentration in transgenic cell lines is essential for more reliable data analysis. Our protocol demonstrates the cluster fluorescence-linked immunosorbent assay (FLISA), a technique that allows the exact quantification of comparable protein expression amounts. Moreover, it enables the detection of clustered or bound subunits of a protein without necessitating ultracentrifugation. In the present protocol, we demonstrate the utilization of two transgene cell lines, each expressing distinct recombinant proteins, to provide comparability of protein yields and detectable subunit clustering.
Key features
• Fast and cost-effective approach to integrate into laboratory practice.
• Different transgene cell lines are comparable regarding their transgene protein expression yields.
• Detection of clustering for up to four different subunits.
• Detection of precise antibody concentration for every protein subunit.
Keywords: Cluster FLISA (Cluster FLISA)Graphical overview
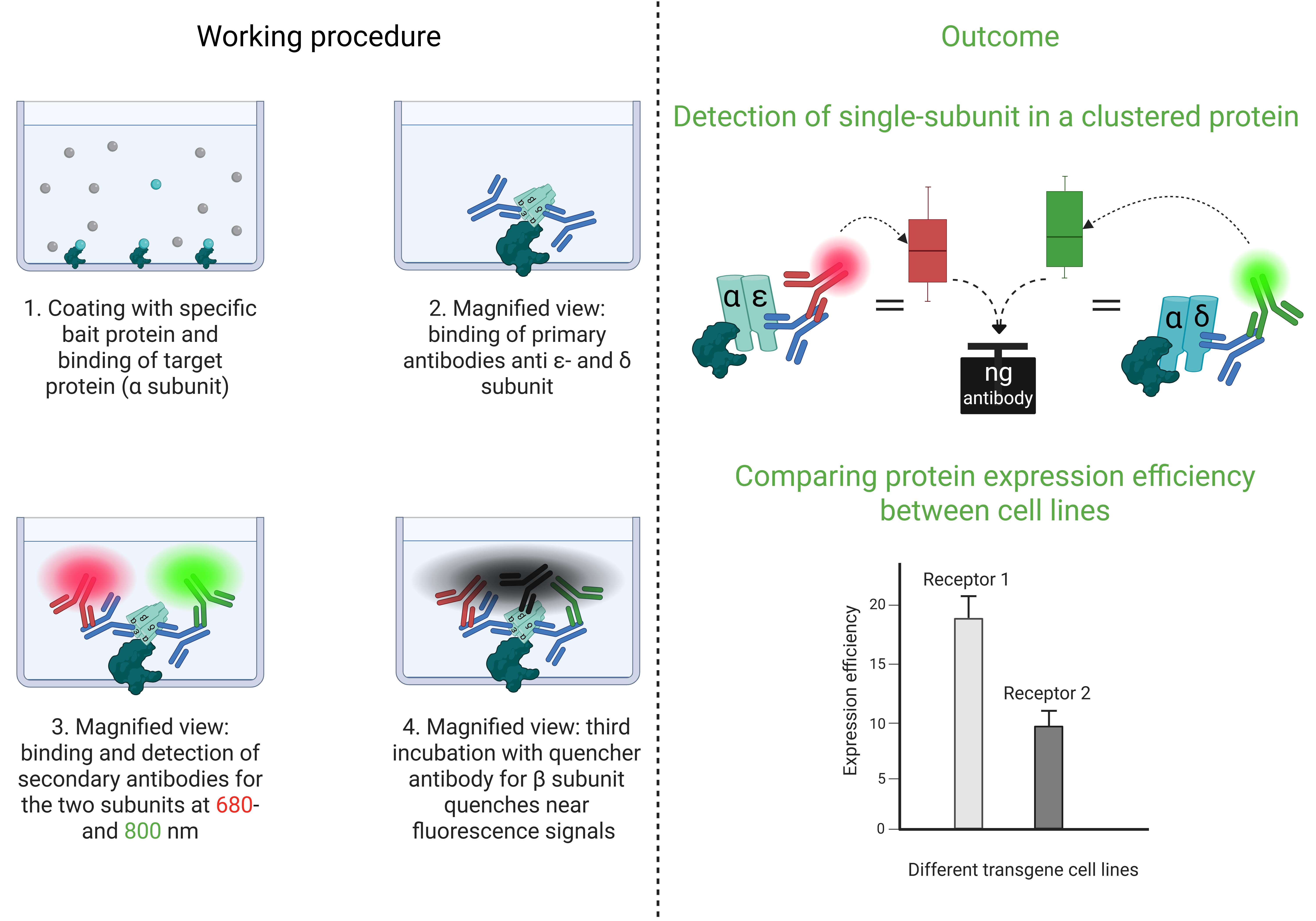
Cluster FLISA procedure and its outcomes
Background
Recombinant cellular model systems are the method of choice for the development of therapeutic approaches in several research laboratories. The generation of numerous transgene cell lines allows for obtaining the most suitable model system for experimental investigations. It is necessary that the cell lines developed exhibit a range of characteristics that can be documented in the results, thereby ensuring the robustness of the data. To this end, we have devised a novel method, the cluster fluorescence-linked immunosorbent assay (cluster FLISA), to systemically validate model systems. We investigated various subtypes of the nicotinic acetylcholine receptor (nAChR), which are exemplarily used in this protocol. In particular, a comparison was made between generated [1,2] and purchased transgene cell lines, in order to determine which cell lines possessed the optimal characteristics for the experimental requirements. The characteristics examined included cellular host viability, the amount of transgene protein, and function. Data comparability was critical to selecting the right cellular system.
The cluster FLISA technique is similar to the known ELISA (enzyme-linked immunosorbent assay) method. The user employs a coated bait protein that binds the target protein [3] and an antibody to determine the concentration [4]. The cluster FLISA method represents an advancement over this method. The procedure is shown in Figure 1. It enables further antibody binding, specifically for each individual subunit of the target protein. By binding to one specific subunit, the bait protein enables the identification of all other subunits in the target complex via their associations. In the case of nAChR cluster determination, it is alpha-bungarotoxin that binds specifically to the α subunit [5–7]. If an additional subunit is detected, both are subjected to clustering or binding. This performance is executed in sequential steps that ensure the individual detection of each target subunit. The use of an antibody labeled with QC-1 quencher allows subsequent investigation of subunit clustering by extinguishing the detected fluorescence signals [8]. Moreover, it enables the use of a detection device with a limited number of fluorescence channels. By normalizing the protein concentration and analyzing the bound antibody concentrations, data can be compared across various cell lines and diverse proteins. The bait protein we used for the positive control of the assay and concentration determination of the bound antibodies was alpha-bungarotoxin conjugated to biotin. Other bait proteins could be made available by determining other target proteins. In addition, cluster FLISA enables the detection of subunit clustering of a protein without the need for ultracentrifugation, which represents a costly investment in the acquisition phase. Analytical ultracentrifugation is a method for determining the size, density, and shape of proteins [9].
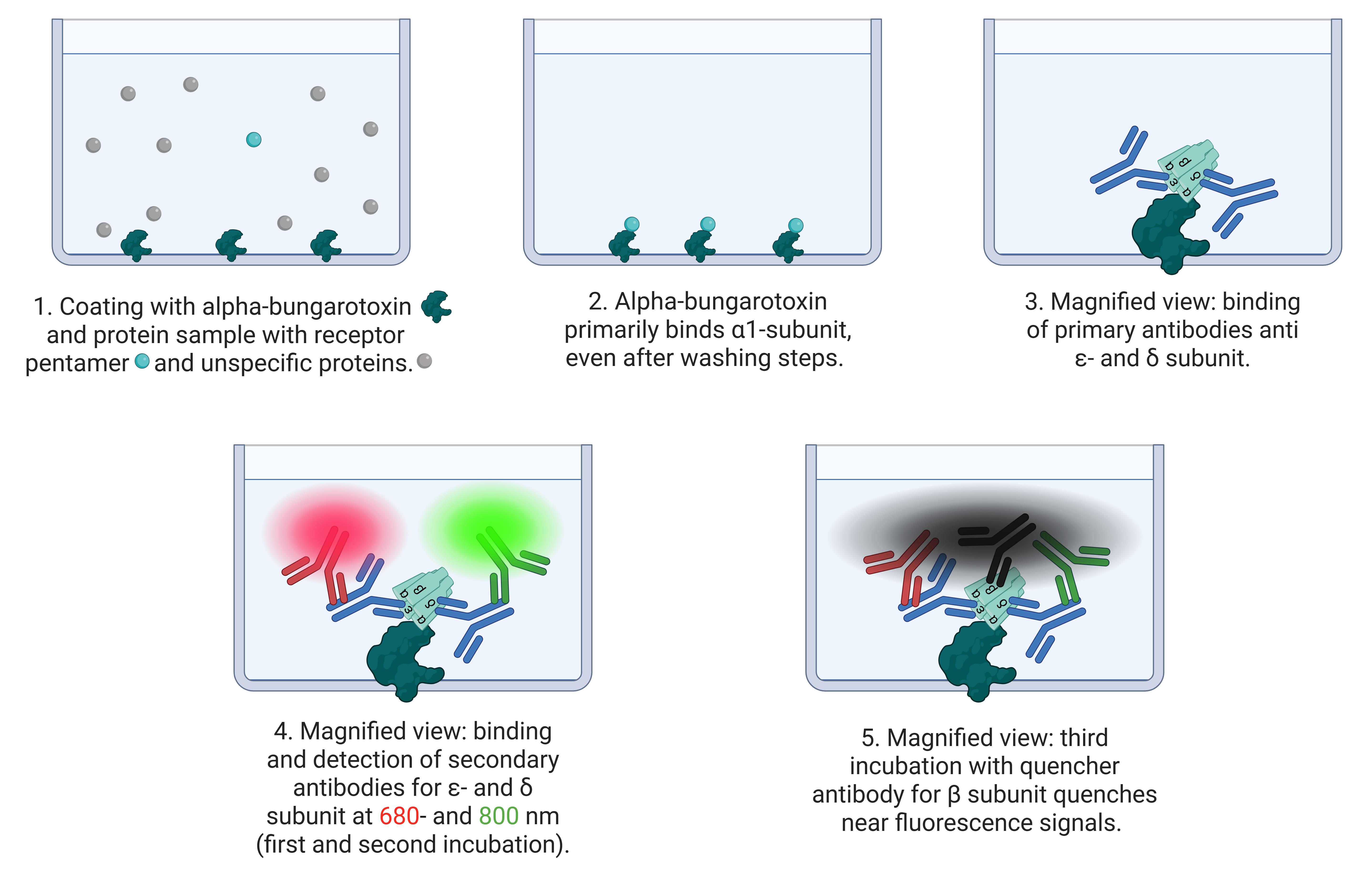
Figure 1. Principle of cluster FLISA assay. The scheme shows the process of cluster FLISA. 1. Coating the 96 microplate wells with the bait protein alpha-bungarotoxin and the protein sample solution. 2. Alpha-bungarotoxin binds receptor pentamers at the α1 subunit and washes out the unspecific protein sample. 3. In different runs, primary antibodies bind to ε- and δ subunits. 4. Subsequent detection by fluorescence-labeled antibodies at 680 and 800 nm. This enables the detection of different individual subunits. 5. In the final run of the cluster FLISA, a quencher-labeled antibody directed against the β1 subunit quenches nearby fluorescent signals. The tightly bound β1 subunit in the target protein is necessary for fluorescence signal quenching.
The concentration of different transgene subunits was determined by measuring the amount of bound antibody. The clustering of the same subunit into a pentameric receptor was also analyzed. Both outcomes provided valuable insights into the quality of the model system. For instance, a transgene cellular model system with a good score for subunit clustering, but a decreased protein concentration, requires improved processing. We hypothesize that the maximum potential of recombinant cellular model systems will be attained by attempting to generate a state of comparable quality. The cluster FLISA is an easy tool for assessing these aspects, and it may be of interest to other research groups to qualify their models.
Materials and reagents
In order to enhance the performance of cluster FLISA, it is necessary for the operator to possess purified transgene proteins. Please note that there are a couple of methods for protein purification, and we do not impose performance requirements for diverse protein columns or magnetic bead-based techniques. However, we recommend determining the protein concentration after the purification procedure to apply similar amounts. The purification of the protein solution used in this protocol was performed as described in our previous publications [1,2]. The subsequent protocol data exemplifies the utilization of cluster FLISA. It describes the exemplary proteins α12β1δε nAChR and α7 nAChR, both of which have a pentameric structure. The α12β1δε nAChR consists of four different subunits in a pentamer, whereas the α7 nAChR is a five-subunit homomeric structured protein. Both transgene cell lines expressing these proteins were generated in our laboratory [1,2]. Operators are required to modify antibodies and bait proteins to their target protein/subunits. Furthermore, we performed our cluster FLISA using the Odyssey CLx (LiCor) for plate scanning, but other devices are also possible for fluorescence scanning. The Odyssey CLx has two detection channels (680 and 800 nm wavelengths), which demonstrate how to use this device or others without scanning channels for every subunit. Any fluorescence scanning device can be used for analysis. Furthermore, the order of subunit detection is not specifically defined and can be freely selected.
Biological materials
Two recombinant Chinese hamster ovary (CHO) cell lines (Leibniz Institute DSMZ, ACC 110) stably expressing the α12β1δε nAChR and the α7 nAChR.
Note: The α12β1δε nAChR consists of genetic tags His(6)-tag for α1, HA-tag for β1, Myc-tag for δ, and FLAG-tag for ε subunit, which was used for unambiguous identical detection.
Reagents
1. ELISA phosphate coating buffer, 10 mM phosphate buffer, pH 7.4 (Invitrogen, Life Technologies, catalog number: CB07100)
2. Blocking buffer (LiCor, catalog number: 927-70001)
3. Bait protein Alpha-bungarotoxin conjugates (Invitrogen, Life Technologies, catalog number: B1601)
Note: The bait protein is target-defined.
4. Bait protein for positive control and concentration determination Alpha-bungarotoxin conjugates Biotin-XX (Invitrogen, Life Technologies, catalog number: B1196)
Note: The bait protein is target-specific.
5. PBS pH 7.4, without Mg2+ and Ca2+ (Gibco, catalog number: 10010-015)
6. Tween 20 (Merck/Sigma-Aldrich, catalog number: P1379-100mL)
7. Primary antibodies: His-tag (Abcam, catalog number: ab18184), HA-tag (Abcam, catalog number: ab49969), myc-tag (Abcam, catalog number: ab32), flag-tag (Abcam, catalog number: ab236777), and α7 subunit (Abcam, catalog number: ab216485)
8. Secondary antibodies: IRDye 800 rabbit (LiCor, catalog number: 92632211), IRDye 800 mouse (LiCor, catalog number: 92632210), IRDye 680 rabbit (LiCor, catalog number: 92668071), IRDye 680 mouse (LiCor, catalog number: 92668070), and IRDye 800 streptavidin antibody (LiCor, catalog number: 92632230)
9. Alexa Fluor dye 680 (Invitrogen, Life Technologies, catalog number: A20188)
Note: Alexa Fluor dye 680 is no longer available; the successor product is Alexa Fluor dye 680, catalog number: A88069. The procedure remains the same.
10. QC-1 quencher dye (LiCor, catalog number: 92970030)
11. Zeba spin desalting columns 7K MWCO, 75 μL (Invitrogen, Life Technologies, catalog number: 89878)
12. Bovine serum albumin (BSA) (Merck/Sigma-Aldrich, catalog number: A7906-50G)
13. Ultra-pure distilled water DNase/RNase-free (Invitrogen by Life Technologies, catalog number: 10977-035)
14. PBST (0.1% Tween-20 in PBS)
15. Primary antibody dilution 1:200 in blocking buffer
Note: Antibody dilution was performed according to the manufacturer's instructions for use with ICC/IF.
16. Secondary antibody dilution 1:1,000 in blocking buffer
Note: Antibody dilution was performed according to the manufacturer's instructions for use with WB.
Solutions
1. Solutions for cluster FLISA and antibody labeling process (see Recipes)
Recipes
1. Solutions for cluster FLISA and antibody labeling process
| Reagent | Final concentration | Notes |
|---|---|---|
| Alpha-bungarotoxin coating buffer | 20 μg/mL | For a sample concentration of ≤ 0.1 mg/mL |
| Alpha-bungarotoxin Biotin-XX coating buffer | 20 μg/mL | For a sample concentration of ≤ 0.1 mg/mL |
| QC-1 solution | 4 μg/μL | The QC-1 dye is based on an NHS-ester formulation, which is unstable at the moment of solvation. |
| PBS-BSA solution | 100 mg/mL | Fresh solution |
Laboratory supplies
1. Black 96-well microplates with clear bottom (Greiner BioOne, catalog number: 655986)
2. pH indicator strips 6.5–10 (Meck, catalog number: 1095430001)
3. Protein LoBind® tube 0.5 mL (Eppendorf, catalog number: 0030108094)
4. Protein LoBind® tube 1.5 mL (Eppendorf, catalog number: 0030108116)
5. Protein LoBind® tube 2 mL (Eppendorf, catalog number: 0030108132)
6. Eppendorf tube screw cap 5 mL (Eppendorf, catalog number: 0030122313)
Equipment
1. Odyssey CLx Imager (LiCor, catalog number: 9140)
Note: Odyssey CLx is no longer available; the successor model is Odyssey DLx Imager, catalog number: 9142.
2. KS 260 control IKA (Sigma-Aldrich, catalog number: Z341835)
3. Fume hood HERA Safe KSP (Thermo Scientific, catalog number: 17168075)
4. Nanodrop 8000 spectrophotometer (Thermo Scientific, model: ND-8000-GL)
5. Mini Spin plus (Eppendorf, catalog number: 5453000011)
6. Rotina 420R (Hettich, catalog number: 4706SET2)
7. Thermo Mixer C (Eppendorf, catalog number: 5382000015)
8. Varioshake VA 15 T (Lauda, catalog number: LDL003059)
Software and datasets
1. Image Studio software (version 5.2 LiCor)
2. Prism [version 9.5.1 (733) GraphPad]
3. Excel (version 16.0.10415.20025, Microsoft Excel 2019 MSO)
4. RStudio (version 2024.09.1 Posit Software, PBC)
5. R (version 4.4.2 R Resources)
Procedure
文章信息
稿件历史记录
提交日期: Jul 30, 2025
接收日期: Sep 24, 2025
在线发布日期: Oct 14, 2025
出版日期: Nov 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Brockmöller, S. and Molitor, L. M. (2025). Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins. Bio-protocol 15(21): e5484. DOI: 10.21769/BioProtoc.5484.
分类
生物化学 > 蛋白质 > 免疫检测
细胞生物学 > 基于细胞的分析方法 > 蛋白合成
生物化学 > 蛋白质 > 表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












