- EN - English
- CN - 中文
Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
利用高效逆转录酶uMRT高效检测环状RNA
发布: 2025年10月20日第15卷第20期 DOI: 10.21769/BioProtoc.5480 浏览次数: 1402
评审: Jibin SadasivanAnu ThomasAnonymous reviewer(s)

相关实验方案

用细胞增殖和基因表达测定法从柏氏血矛线虫排泄物和分泌物中筛选具有免疫潜力的分子
Jocelyn Maza-Lopez [...] Carla O. Contreras-Ochoa
2023年06月20日 1329 阅读
Abstract
Circular RNAs (circRNAs) are covalently closed RNA molecules known for their increased stability compared to linear RNAs. Synthetic circRNAs are being developed as RNA therapeutics, while natural circRNAs are being investigated for their biological roles in eukaryotes and their potential as disease biomarkers. Consequently, the accurate detection and validation of circRNAs is crucial for advancements in both fundamental RNA research and biotechnological applications. Common methods for circRNA validation involve RT-PCR using divergent primers, followed by sequencing across the circRNA junction. However, most described methods are high-throughput approaches that require time-consuming RNA processing steps, and they are unable to detect highly structured circRNAs. Additionally, methods for low-throughput sequencing of small circRNAs (<150 nt) require cloning prior to sequencing. A simplified protocol for the validation of circRNA sequences irrespective of structure, sequence complexity, and length has not yet been described. In this method, we describe an improved RT-PCR protocol for circRNA detection by using UltraMarathonRT® (uMRT), a highly processive reverse transcriptase. Unlike other reverse transcriptases, uMRT can reverse-transcribe large, structured circRNAs of varying sizes, at ambient temperatures, enabling sequencing of the resulting concatemeric amplicons generated by RT-PCR and other methods. Using this method, we sequenced circRNAs containing highly structured internal ribosome entry sites commonly utilized in synthetic circRNAs, natural circRNAs containing repetitive elements, and small circRNAs, all without the need for cloning. With this new platform, we offer a protocol for the precise detection of nearly any circRNA species.
Key features
• This protocol shortens current methods for circRNA detection and sequencing by sequencing RT-PCR products directly, without the need for cloning or processing the PCR product.
• This protocol describes a simple and cost-effective RT-PCR method for single circRNAs.
• The highly processive UltraMarathonRT (uMRT) functions at ambient conditions, reducing circRNA degradation.
• This protocol enables sequencing of circRNAs that are below 150 nt as well as larger circRNAs.
• This protocol enables the detection of structured and very large circRNAs.
Keywords: circRNA (环状RNA(circRNA))Graphical overview
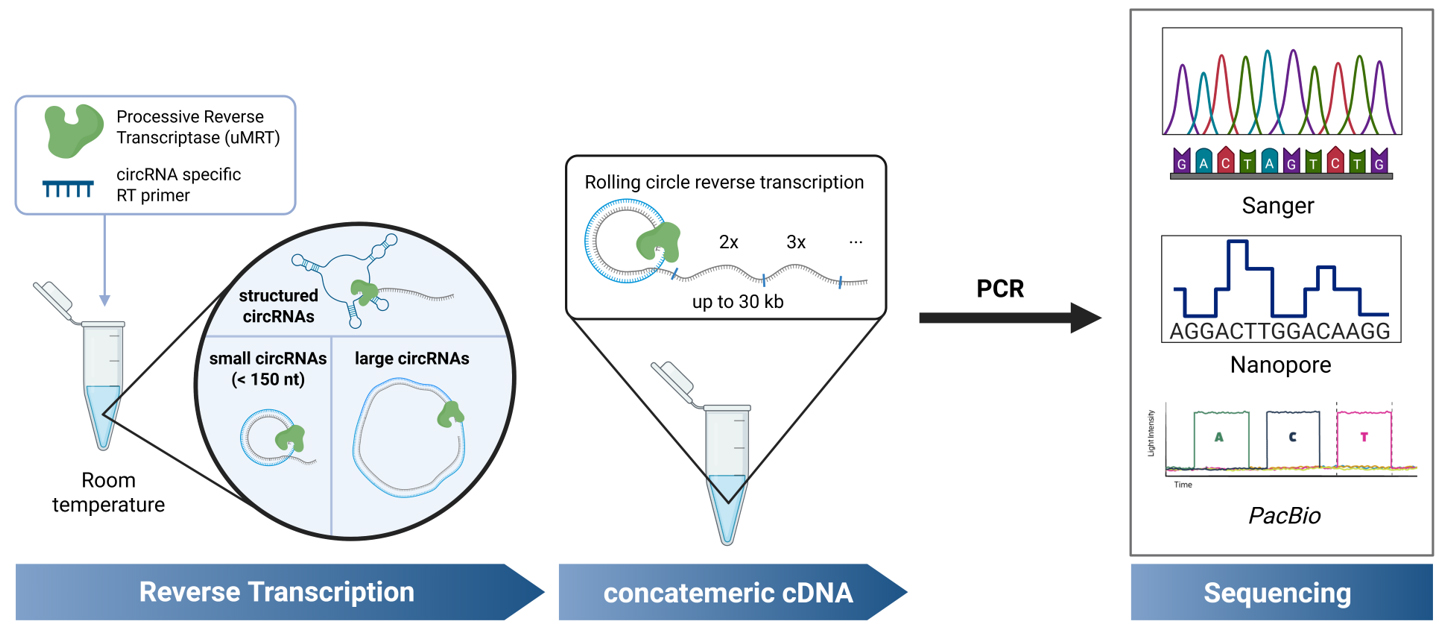
Background
Circular RNAs (circRNAs) are covalently closed RNA molecules that exhibit increased stability due to their resistance to exonuclease cleavage [1]. circRNAs have been implicated in a variety of biological functions [2], including human diseases such as cancer [3] and neurological disorders [4], where circRNAs have shown potential as biomarkers [5]. Additionally, circRNAs are considered attractive candidates for RNA therapeutics because of their enhanced stability and protein production compared to linear RNAs [6,7]. Given the growing interest in circRNAs for these applications, accurate detection and sequence validation are crucial.
Numerous RT-PCR-based sequencing methods have been described for identifying natural circRNAs in cell extracts [8,9]. These are high-throughput methods designed to detect the circRNA transcriptome by utilizing RNA enrichment strategies, such as ribosomal RNA (rRNA) depletion and RNase R digestion (to eliminate linear RNAs) [10]. However, these approaches are too costly and time-consuming for the verification of individual circRNAs, such as those synthetically made or individual natural circRNAs. Currently, individual circRNAs are sequence-verified through RT-PCR followed by Sanger sequencing across the circRNA junction, or nanopore sequencing [6,7,11,12]. However, these approaches have several notable limitations. For small circRNAs (~100 nt), the resulting RT-PCR product is too short for Sanger and Nanopore sequencing, thus necessitating a cloning step prior to sequencing, as utilized by Du et al. [7]. This additional cloning step increases the time and cost of the verification process. Additionally, RT-PCR is typically performed using retroviral reverse transcriptases (RTs) [13,14], which stall on structured RNAs [15], restricting detection to unstructured circRNAs within a limited size window. While more highly processive RTs have been employed for circRNA detection [16], existing protocols require the use of high temperatures during cDNA synthesis that may shear and degrade RNA circles. Moreover, they have only been used for high-throughput sequencing of cellular circRNAs. Currently, there is no simple protocol for sequencing individual synthetic or cellular circRNA sequences by RT-PCR, irrespective of circRNA structure, sequence complexity, or length.
The method described here enables the straightforward validation of individual circRNA sequences, without the need for enrichment strategies or cloning, by using the highly processive UltraMarathonRT (uMRT) [17,18]. This approach utilizes RT-PCR with uMRT and commercial Nanopore or Sanger sequencing to rapidly and cost-effectively validate small, large, and structured circular RNAs at room temperature. Although this protocol focuses on validation of individual circRNAs, these highly processive RTs could be used for transcriptome-wide RNA sequencing of natural circRNAs or in sequencing approaches that circularize a transcriptome [19]. Applying uMRT to transcriptome-level approaches will require further experiments, but it has the potential to reveal novel circRNAs, including those containing structured regions and other complex elements.
Materials and reagents
Reagents
1. DNA Clean and Concentrator-25 (Zymo Research, catalog number: D4033)
2. uMRT Reverse Transcriptase kit (RNAConnect, catalog number: catalog number: R1002S)
3. Deoxynucleotide (dNTP) solution mix (10 mM each) (NEB, catalog number: N0447S)
4. RNaseOUTTM (InvitrogenTM, catalog number: 10777019)
5. CircRNA template or cell extracted RNA
6. RNA loading dye (2×) (NEB, catalog number: B0363S)
7. Reverse transcription primer (should not span circRNA junction)
8. DreamTaq PCR Master Mix (Thermo Fisher Scientific, catalog number: K1081)
9. Divergent PCR primer pair (unique to the circRNA)
10. Sodium chloride (NaCl) (Millipore Sigma, catalog number: S9888)
11. MOPS, sodium salt (Na-MOPS) (Research Products International, catalog number: M92040)
12. Trizma® base (Millipore Sigma, catalog number: T1503)
13. EDTA (0.5 M) (Invitrogen, catalog number: AM9260G)
14. Glacial acetic acid (Millipore Sigma, catalog number: A6283)
15. SeaKem® LE agarose (Thermo Fisher Scientific, catalog number: BMA50001)
16. GelRed® nucleic acid gel stain (Biotium, catalog number: 41003)
17. GeneRuler 1 kb Plus DNA ladder (Thermo Fisher Scientific, catalog number: SM1331)
18. TRIzol reagent (Thermo Fisher Scientific, catalog number: 15596026)
19. RNase R (NEB, catalog number: M0100S)
20. RNA Clean and Concentrator-25 (Zymo Research, catalog number: R1017)
21. Nuclease-free water (NEB, catalog number: B1500S)
Solutions
1. RNA elution buffer (see Recipes)
2. 10× TAE (see Recipes)
Recipes
1. RNA elution buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| 1 M Na-MOPS (pH 6.0) | 10 mM | 200 μL |
| 5 M NaCl | 300 mM | 1200 μL |
| 0.5 M EDTA (pH 8.0) | 1 mM | 40 μL |
| Deionized H2O | n/a | 18.56 mL |
| Total | n/a | 20 mL |
2. 10× TAE
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Trizma® base | 400 mM | 48.5 g |
| 17.4M Glacial acetic acid | 200 mM | 11.4 mL |
| 0.5 M EDTA (pH 8.0) | 10 mM | 20 mL |
| Deionized H2O | n/a | Fill up to 1 L |
| Total | 1 L |
Laboratory supplies
1. Standard pipette tips with a volume capacity of 10 μL, 20 μL, 200 μL, and 1 mL
2. PCR strip tubes (USA Scientific, catalog number: 1402-4708)
3. 1.7 mL tubes (Genesee, catalog number: 24-282)
4. 0.22 μm syringe filters (Millipore Sigma, catalog number: WHA9913-2502)
5. 1 mL disposable syringe (Thermo Fisher Scientific, catalog number: 14-823-30)
6. Tissue culture flask, 75 cm2 (VWR, catalog number: 13-680-65)
7. Serological pipettes 10 mL (Genesee, catalog number: 12-104)
Equipment
1. Pipetman (2, 20, 200, 1,000 μL) (Gilson, catalog number: F167360)
2. EppendorfTM Centrifuge 5425 (Eppendorf, catalog number: 5405000247)
3. C1000 Touch Thermal Cycler (Bio-Rad Laboratories, catalog number: 1851148)
4. Sub-Cell GT Horizontal Electrophoresis System (Bio-Rad Laboratories, catalog number: 1704403)
5. GelDoc Go (Bio-Rad Laboratories, catalog number: 12009077)
6. Nanodrop 1000 (Thermo ScientificTM)
7. Personal microcentrifuge (USA Scientific, catalog number: 2641-0016)
Software and datasets
1. Benchling (Benchling, Non-Validated Cloud)
This is a free-to-use cloud software. The sequence alignment tool in Benchling was used for the analysis of the Sanger and Nanopore sequencing data.
Procedure
文章信息
稿件历史记录
提交日期: Jul 30, 2025
接收日期: Sep 17, 2025
在线发布日期: Sep 26, 2025
出版日期: Oct 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Warkentin, R. and Pyle, A. M. (2025). Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT. Bio-protocol 15(20): e5480. DOI: 10.21769/BioProtoc.5480.
分类
分子生物学 > RNA > RNA 检测
分子生物学 > RNA > qRT-PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










